 Research Article
Research Article
Investigation of Leptin Receptor and eNOS Gene Polymorphisms in Preeclamptic Umbilical Cords
Fatma Secer Celik1*, Esra Celen1,4, Hatice Kubra Yildiz1, Safaa Altweish1, Huseyin Donmez1, Yasemin Gurbuz1, Ayten Demirci2, Esra Buyuk2, Esra Un Arslan,2, Merve Ilhan3, Ayse Saide Sahin2 and Hasibe Vural1
1Department of Medical Biology, Necmettin Erbakan University, Turkey
2Department of Medical Pharmacology, Necmettin Erbakan University, Turkey
3Department of Medical Biochemistry, Necmettin Erbakan University, Turkey
4Faculty of Agriculture and Natural Sciences, Konya Food and Agriculture University, Turkey
Fatma Secer Celik, Necmettin Erbakan University, MerMedical Faculty, Department of Medical Biology, Turkey.
Received Date: November 27, 2018; Published Date: December 06, 2018
Abstract
Background: Preeclampsia occurs in the umbilical artery with proteinuria and edema accompanying pregnancy-induced hypertension. Vasoconstriction results in severe complications such as intrauterine growth restriction, preterm birth, fetal death due to decreased uteroplacental blood flow. Preeclampsia may cause perinatal mortality and morbidity and it is one of the most important causes of maternal mortality. The etiology and pathogenesis of preeclampsia, which is present in 7,18% of all pregnancies, has not yet been elucidated. However, it is thought that one of the causes of pathogenesis may be caused by possible changes in the synthesis of endothelial nitric oxide synthase (eNOS) mediated nitric oxide (NO). DNA sequencing studies revealed many single nucleotide polymorphisms (SNPs) in the promoter region, exons and introns of this gene. T-786C SNP is found in the promoter region of the eNOS gene. There are several agents affecting eNOS activity in preeclampsia. Leptin is one of these agents. The exact reason of why leptin, which is a vasodilator agent, has no effect on preeclamptic pregnancies, is not known yet. However, it is thought that the leptin receptor gene Lys109Arg SNP in exon4 and Gly223Arg SNP in exon6 may affect receptor function and signaling.
Materials and methods: We investigated polymorphisms of eNOS and leptin receptor genes with HRM and PCR-RFLP techniques.
Results: Both analyses showed no SNPs in exon4 of leptin receptor (LEPR) gene in preeclampsia patients suggesting that there should be no relationship with exon4 of LEPR gene and preeclampsia. We found that there were SNPs in promoter region of eNOS gene and exon6 of LEPR gene.
Conclusion: HRM and PCR-RFLP gave different number of SNPs, these SNPs of these genes could be related with preeclampsia. This study is a preliminary step for our next molecular studies although few numbers of the samples were included. Increasing the number of samples would give the statistically significant results.
Keywords: Leptin, Preeclampsia, Polymorphism
Introduction
Preeclampsia is associated with high blood pressure and proteinuria during pregnancy. Preeclampsia causes complications in 2-8% of all pregnancies, premature birth in 15% and maternal death in 9-26% of all pregnancies [1]. In preeclampsia patients, decrease in uteroplacental blood flow rate as a result of vasoconstriction in the umbilical artery may have serious consequences such as intrauterine growth restriction, preterm birth, fetal death. The etiology of preeclampsia is not clarified yet. However, it is thought that lipid peroxidation due to the activity of reactive oxygen species (ROS) may be the reason. When increased oxygen demand cannot be met during pregnancy, the increase of oxidative stress factors causes cell damage in fetus due to lipid peroxidation and decrease in perfusion as a result of vasoconstriction in the umbilical artery [2].
Vasoconstriction occurs by increasing the intracellular Ca2+ concentration via the Ca2+ channel [3]. However, there are some agents that cause vasodilatation in vascular smooth muscle. One of these agents is leptin, which causes nitric oxide (NO) mediated vasodilatation [4]. Leptin which is an adipokine hormone and leptin receptors are more than adipose tissue in the umbilical cord. This situation suggests that there should be an important function of leptin and leptin receptors in pregnancy. Several mutations have been identified in the leptin receptor gene. Any mutation in the leptin receptor gene may result in a defective signal transduction. Leptin in the target tissue could not fully perform its task. Most of these mutations result in early onset of obesity, hyperphagia and infertility [5].
The NOS enzyme catalyzes the NO formation reaction as a result of the interaction of L-arginine with the terminal guanidino nitrogen group and molecular oxygen. The eNOS and nNOS isoforms are active when the calcium-calmodulin complex in the cell is bound to the enzyme. Therefore, NOS activity is associated with intracellular calcium level. On the other hand, iNOS always has a firmly bound calmodulin. Therefore, the activity of the iNOS isoform is independent of the calcium level and the enzyme is always in a position to be active [6].
The R223Q polymorphism (rs1137101), one of the LEPR polymorphisms, is formed by the change of arginine to glutamine in codon 223 of exon6. There is a variety of information in the literature about the link between obesity and this polymorphism [7]. Leptin sensitivity in humans is thought to be related to variations in leptin or leptin receptor genes. Polymorphism in leptin receptor genes has not been shown to be associated with hypertension but leptin gene polymorphism has been reported to be associated with hypertension risk [8]. Another LEPR polymorphism is the R109K (rs1137100) polymorphism in codon 109 of exon 4. There are studies showing that obesity and sugar preference are associated with genetic variations. The first report of these was shown by Mizuta et al. [9]. In this study, LEPR Arg223Gln, LEPR Arg109Lys and eNOS -786T>C polymorphisms were investigated in normal and preeclamptic cord tissues.
Materials and Methods
Samples
Normal and preeclamptic pregnant women between the ages of 18-45 gave birth at in Necmettin Erbakan University, Meram Faculty of Medicine, Department of Obstetrics Gynecology (Medical Faculty Dean’s Office Ethics Committee approval number is 2018/1273). After delivery, 10-15 cm sections were taken from umbilical cords and put directly into cold Krebs-Henseleit (KHS) solution (NaCl: 119mM, KCl: 4,7mM, MgSO4.7H2O: 1,5mM, KH2PO4: 1,2mM, CaCl2.6H2O: 2,5mM, NaHCO3: 25mM, Glucose: 11Mm). The umbilical cord samples were stored at +4 °C until used for the experiments. DNA samples isolated from healthy individuals were used as positive control samples.
DNA isolation
The tissues were homogenized with liquid nitrogen. Homogenized tissues were filled with 2X lysis buffer (10X Lysis buffer: 770mM NH4Cl, 10mM EDTA, 46mM KHCO3) up to 15ml and were thoroughly mixed for 10 minutes. The tubes were kept on ice for 30 minutes and then centrifuged at 3000 rpm for 10 minutes at +4ºC. The supernatant phase was discarded. 0,75ml of SALT/EDTA (75mM NaCl, 25 mM EDTA) was added onto pellet and vortexed thoroughly. Then, 0,075ml 10% SDS solution, 37,5μl Proteinase K (10mg/ml) was added and the samples were incubated at 55 °C for 3 hours. 0,75ml phenol (pH 8.0) was added into the tube. The mixture was mixed for 20 seconds and then inverted for 5 minutes. The tubes were centrifuged at 3000rpm for 10min at +4 °C. After centrifugation, the supernatant phase was taken into new sterile tubes and 0,75 ml phenol chloroform: isoamyl alcohol (25:24:1) was added. The mixture was mixed for 20 seconds and then inverted for 5 minutes. The tubes were centrifuged at 3000rpm for 10 min at +4 °C. At the end of the centrifugation, supernatant phase in the tubes was taken into new tubes and 1/10 of NaAc were added to the supernatant. 95% ethanol was added twice as much as the current volume. The precipitated DNA was taken into the new tube with the glass rod. The pellet was dried to remove the alcohol. The pellet was diluted with distilled water before use.
High resolution melting (HRM) curve analysis
The real-time PCR-based HRM technique was used to detect the polymorphisms of the interested genes. Briefly, 50ng DNA samples isolated from each group, 10μl of 2X HRM mixture, 50μl forward primer, 50μl reverse primer and dH2O until 20μl were mixed. The reaction mixtures were placed on the PCR System/BioRad CFX Connect and the HRM protocol consisting of denaturation, amplification and melting steps were applied. The denaturation stage was carried out at 95 °C for 10 minutes and the temperature required for the primers to attach to the target site was chosen as 60 °C. The polymerization of the complementary DNA was performed at 72 °C. The cycle consisting of successive application of these three different temperatures was repeated for 40 times. Following PCR amplification, high-resolution melting was performed: heated to 95 °C for 30 seconds and then cooled to 60 °C for 1 minute and performed from 65 °C to 95 °C, rising at 0.2 °C/s. The graphs were analyzed in the BioRad Precision Melt Analysis program to analyze the possible polymorphisms. In this study, primers belonging to the exon6, exon4 of LEPR gene and the promoter region of eNOS gene are shown in (Tables 1 & 2).
Table 1: Primers and PCR product lengths of the promoter region of the exon6, exon4 and eNOS gene of the OBRb gene.
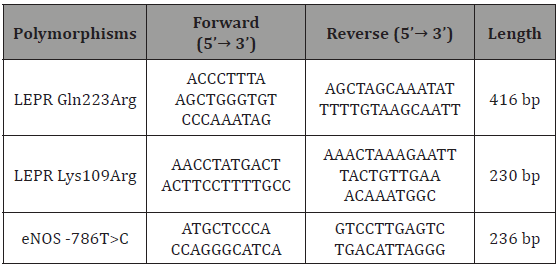
Table 2: Comparison of the results of HRM and PCR-RFLP analysis.
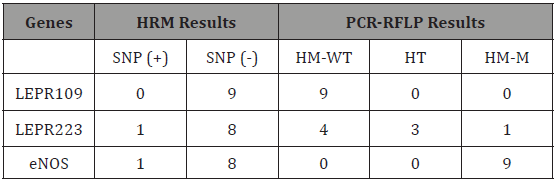
WT: wild type, MT: mutant type, HT: heterozygous type, HM: homozygous type.
PCR-RFLP analysis
For polymorphism analysis with PCR-RFLP, the target gene sequences were amplified by Polymerase Chain Reaction (PCR) using primers which were also used in HRM analysis to cover each target mutation. All PCR reactions were run with 50ng of DNA and 25μl of PCR mix (1X PCR buffer, 200μM dNTP, 1,5 mM MgCl2, 0,025 U Taq DNA polymerase, 5 pmol forward and reverse primer). Samples were runned to 35 cycles of PCR protocol for 5 minutes at 95 °C for pre-denaturation, 30sec at 94 °C, 30 seconds at appropriate primer binding temperature and 30 seconds at 72 °C. Then PCR samples were run on agarose 1% gel and genotyped by RFLP technique using the MspI and HaeIII restriction enzymes for each mutation. For this purpose, 10μl PCR sample was cut at appropriate temperature with the appropriate restriction endonuclease enzyme covering each mutation site. Then, the samples were run on 2% agarose gel and genotyping was performed.
Results
HRM analysis results
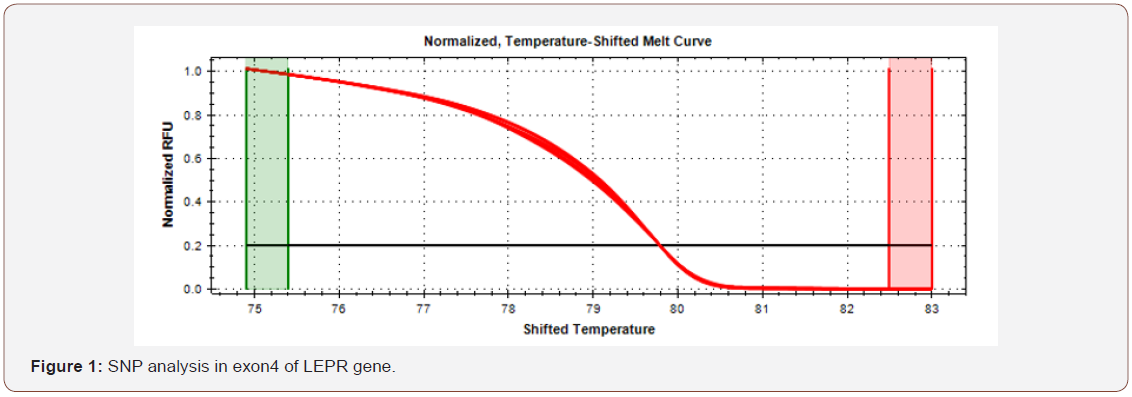
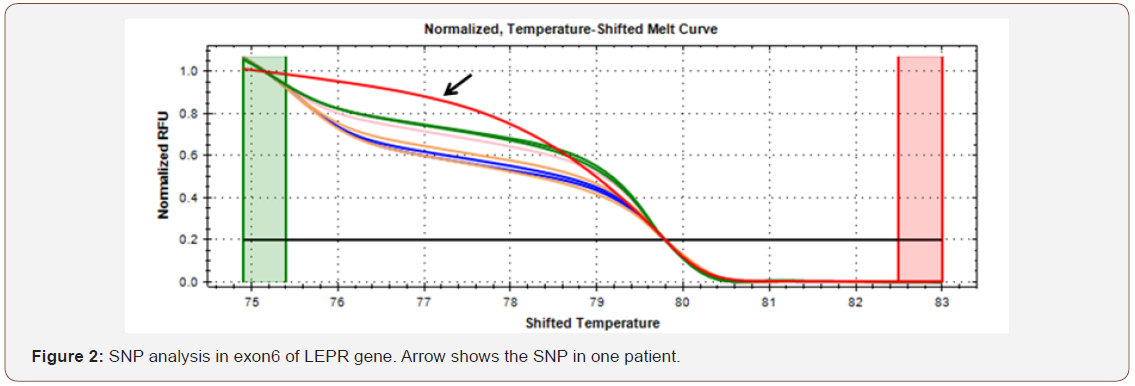
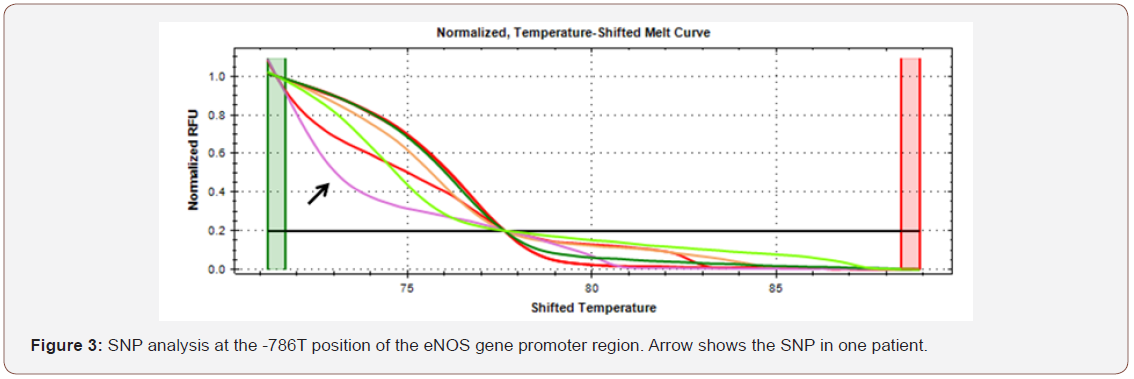
HRM results showed that there was no change in A>G in exon4 of LEPR gene (Figure 1). In exon6 of LEPR gene, SNP was detected in one patient (Figure 2). The red arrow shows the SNP in that patient. The purple arrow in Figure 3 shows that there was SNP in one patient which is C>T change in the promoter region of eNOS gene.
PCR-RFLP analysis results
The lengths of the regions synthesized by specific primers of the polymorphism of rs1137101 found in LEPR exon6 was determined to be 416bp. All of the samples obtained from 3 positive controls, 3 healthy cord and 3 preeclamptic cord tissues successfully. Then, SNP analysis was performed by the restriction enzyme digestion which we used specifically for this region. As a result, one healthy cord tissue, one preeclamptic cord tissue and one control sample were digested by restriction enzyme. So, these samples were heterozygous for that region. One positive control sample was also digested and it was homozygous for that region as shown in Figures 4-7. The sizes of fragments as heterozygous is 416bp, 292bp and 124bp. The sizes of homozygous fragments are 295bp and 124bp.
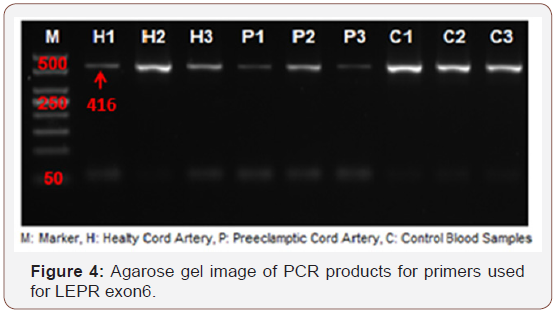
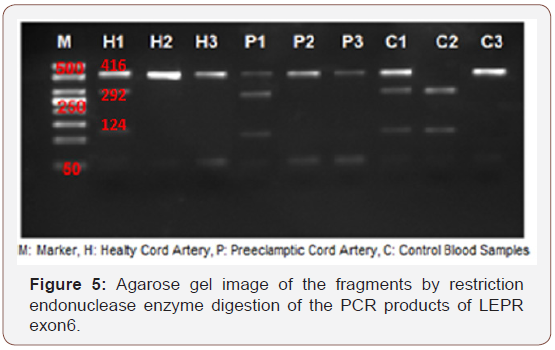
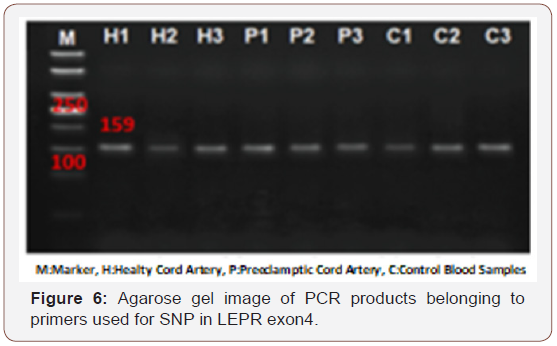
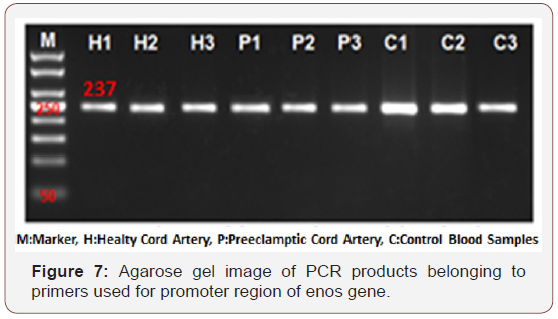
The length of the PCR product for the primers used for SNP in exon4 of LEPR gene was found to be 159bp. Then the result of the restriction enzyme digestion, samples were uncut. The length of the PCR product for the primers used for SNP in the promoter region of eNOS gene was 237bp. As a result of the restriction enzyme digestion, no sample was cut, so we suggested that there was no SNP in this gene.
Discussion
In this study, we isolated DNA samples from 3 healthy cord tissues, 3 preeclamptic cord tissues and 3 healthy blood sample as positive control and investigated the polymorphisms in leptin receptor and eNOS genes. In order to evaluate the polymorphisms, we used HRM and PCR-RFLP techniques. HRM analysis was used because it is relatively affordable, rapid and sensitive method. In literature, there are many studies which used HRM technique for SNP analysis [10]. In this study, HRM analysis was performed in order to validate the PCR-RFLP results. In the result of HRM analysis, different genotypes give different denaturation patterns. Because of different homo- and heteroduplex forms complex curves are formed. Homozygous and heterozygous samples could be differentiated by this method [11]. The results of HRM analysis and PCR-RFLP showed different mutation patterns. This difference could be originated from sensitivity degrees of the methods. HRM analysis is more sensitive than PCR-RFLP method and PCR-RFLP method have more steps to be optimized.
HRM results suggest that SNPs in eNOS gene promoter and exon6 of LEPR could be related with preeclampsia. There was no SNP detected in exon4 of leptin receptor gene in preeclampsia patients. This result shows that this SNP is not related with preeclampsia. However, our sample number is not sufficient to suggest this relationship. The increase of the sample number will provide better understanding about the relationship between these SNPs and preeclampsia.
We detected LEPR Gln223Arg polymorphism but no LEPR Lys109Arg polymorphism in our study. Yang et al. showed that there is not a relationship between LEP G2548A and LEPR Gln223Arg polymorphisms in patients with gestational diabetes and normal glucose tolerance [12]. A meta-analysis investigated the relationship between LEPR gene polymorphisms and hypertension risk included a total of 5955 patients with hypertension and 3830 healthy controls. Accordingly, Gln223Arg polymorphism was found to be significantly higher in hypertensive patients than the control group but there was no statistically significant difference in Lys109Arg polymorphism [13]. In another study in North China, Liu et al. investigated whether the Lys109arg, Gln223Arg and Lys656Asn polymorphisms of LEPR gene have a role in the susceptibility to essential hypertension (EH). They found that there was no significant relationship between Lys109Arg or Lys656Asn polymorphism and EH risk. However, LEPR Gln223Arg polymorphism has an important role in susceptibility to EH [14]. Pena et al. stated that the association of Gln223Arg polymorphism with hypertension was not determined [15].
Polymorphism was detected in the promoter of the eNOS gene in our study. In an Iranian study, there is no relationship was found between eNOS and ACE gene polymorphisms and gestational diabetes among pregnant women in the Iranian population when they were investigated whether there is a relationship between this factor [16].
Conclusion
This study is a preliminary step for our next molecular studies although few numbers of the samples were included. Increasing the number of samples would give the statistically significant results. In addition to the present polymorphism analyses, we are planning to investigate the effect of the LEPR (Leptin Receptor) gene, JAK2 and STAT3 genes expression in preeclamptic pregnant women in our next research.
Acknowledgement
None.
Conflict of interest
No Conflict of Interest.
References
- Magee LA, Pels A, Helewa M, Rey E, von Dadelszen P, et al. (2014) Canadian Hypertensive Disorders of Pregnancy (HDP) Working Group. Diagnosis, evaluation, and management of the hypertensive disorders of pregnancy. Pregnancy Hypertens 4(2): 105-145.
- Sağol S, Özkınay E (2000) Lipid peroxidation in the etiopathogenesis of preeclampsia. T Klin J Gynecol Obst 10: 7-15.
- Karaki H, Ozaki H, Hori M, Mitsui-Saito M, Amano K, et al. (1997) Calcium movements, distribution, and functions in smooth muscle. Pharmacol Rev 49(2): 157-230.
- Rahmouni K, Haynes WG (2005) Endothelial effects of leptin: implications in health and diseases. Curr Diab Rep 5(4): 260-266.
- Israel D, Chua Jr S (2010) Leptin receptor modulation of adiposity and fertility. Trends in Endocrinology & Metabolism, 21(1), 10-16.
- Huang PL (2009)eNOS, metabolic syndrome and cardiovascular disease. Trends Endocrinol Metab 20(6): 295-302.
- Ghalandari H, Hosseini-Esfahani F, Mirmiran P (2015) The association of polymorphisms in leptin/leptin receptor genes and ghrelin/ghrelin receptor genes with overweight/obesity and the related metabolic disturbances: A review. Int J Endocrinol Metab 13(3): e19073.
- Vitoratos N, Chrystodoulacos G, Kouskouni E, Salamalekis E, Creatsas G (2001) Alterations of maternal and fetal leptin concentrations in hypertensive disorders of pregnancy. Eur J Obstet Gynecol Reprod Biol 96(1): 59-62.
- Mizuta E, Kokubo Y, Yamanaka I, Miyamoto Y, Okayama A, et al. (2008) Leptin gene and leptin receptor gene polymorphisms are associated with sweet preference and obesity. Hypertens Res 31(6): 1069-1077.
- Kuzpınar A (2007) High resolution melting curve (HRM) analysis technique for detection of deletion type mutations. Hacettepe University Institute of Health Sciences, MSc. Thesis in Medical Biology, Ankara.
- Twist GP, Gaedigk R, Leeder JS, Gaedigk A (2013) High-resolution melt analysis to detect sequence variations in highly homologous gene regions: application to CYP2B6. Pharmacogenomics 14(8): 913-922.
- Yang M, Peng S, Li W, Wan Z, Fan L, et al. (2016) Relationships between plasma leptin levels, leptin G2548A, leptin receptor Gln223Arg polymorphisms and gestational diabetes mellitus in Chinese population. Scientific reports 6: 23948.
- Lian Y, Tang Z, Xie Y, Chen Z (2015) Leptin receptor gene polymorphisms and risk of hypertension: a meta-analysis. Int J Clin Exp Med 8(8): 14277-14282
- Liu Y, Lou YQ, Liu K, Liu JL, Wang ZG, et al. (2014) Role of leptin receptor gene polymorphisms in susceptibility to the development of essential hypertension: a case-control association study in a Northern Han Chinese population. J Hum Hypertens 28(9): 551-556.
- Graças Pena GD, Guimarães AL, Veloso RR, Reis TC, Gomes CS, et al. (2014) Leptin receptor gene Gln223Arg polymorphism is not associated with hypertension: a preliminary population-based cross-sectional study. Cardiology Research and Practice 2014: 1-7.
- Mirfeizi M, Hasanzad M, Sattari M, Afshari M, Abbasi D, et al. (2018) Association of eNOS and ACE gene polymorphisms as a genetic risk factor in gestational diabetes in Iranian women. Journal of Diabetes and Metabolic Disorders: 1-5.
-
Fatma Secer Celik, Esra Celen, Hatice Kubra Yildiz, Safaa Altweish, Huseyin Donmez, et al. Investigation of Leptin Receptor and eNOS Gene Polymorphisms in Preeclamptic Umbilical Cords. W J Gynecol Women’s Health. 1(3): 2018. WJGWH.MS.ID.000515.
-
Leptin, Preeclampsia, Polymorphism, Enos Gene, Umbilical Cords, Proteinuria, Pregnancy, Hypertension, Vasoconstriction, Intrauterine, Preterm birth, Fetal death, Uteroplacental, Perinatal, Mortality, Morbidity, Vasodilator, Reactive oxygen species, Vasodilatation, Adipokine hormone.
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






