Research Article
Research Article
Localization of Herpetic Viruses Patterns in Sinonasal and Nasopharyngeal Malignant and Benign Tumors
Marwa Mohammed Ali Jassim1, Majid Mohammed Mahmood1* and Saad Hasan Mohammed Ali2
1Department of Biology, Mustansiriyah University, Iraq
2Diseases Research Unit, University of Baghdad, Iraq
Majid Mohammed Mahmood, Department of Biology, College of Science, Mustansiriyah university, Baghdad, Iraq.
Received Date: August 31, 2019; Published Date: September 16, 2019
Abstract
Background: Sinonasal and Nasopharyngeal carcinomas are related to many environmental and genetic predisposing factors. Epstein Barr viruses are well known to induce tumorigenesis in proliferative epithelial tissues. HCMV plays an “oncomodulatory” role in the neoplastic process and can induce cellular responses that would provide a growth advantage for neoplastic cells.
Objectives: This study designed to determine the herpetic viruses’ patterns (punctuated, diffused and mixed) in tissues from Nasopharyngeal and sinonasal lesions.
Methods: The study included a total number of (183) formalin fixed, paraffin-embedded tissue blocks including 35 inflammatory nasal polyps (INP), 35 sinonasal papillomas (SNP), 65 nasopharyngeal carcinomas (NPC), 18 sinonasal carcinomas (SNC) as well as 30 nasal healthy tissues as control group. After histopathological confirmatory re-examination, a Chromogenic in Situ Hybridization technique (CISH) for Epstein Barr virus encoded RNA (EBV-EBERs) and Human cytomegalovirus phosphoproteins 65 (HCMV-PP65) localization were performed.
Results: The mean (20.43) of EBV punctuators among tissues with malignant nasopharyngeal carcinoma was higher than the mean of punctuators (17.00) in sinonasal carcinoma group, whereas the mean of diffused form (71.89) in malignant sinonasal carcinoma tissues was higher than the mean (66.11) in nasopharyngeal carcinoma tissues. On the other hand, the mean of punctuated pattern in malignant sinonasal carcinoma tissues infected with HCMV was higher (21.56) than the means (16.94) of this pattern in nasopharyngeal carcinoma tissues, as well as the mean of diffused pattern in tissues with sinonasal carcinoma was higher (45.11) than its mean in HCMV infected nasopharyngeal carcinoma tissues (34.881). Conclusion: The present findings indicate the impact of both punctuated and diffused patterns in sinonasal and nasopharyngeal pathogenesis and carcinogenesis.
Keywords: NPC; SNC; EBV-EBERS; HCMV-PP65; CISH
Introduction
Nasopharyngeal cancer is a highly malignant tumor, the primary tumor mass can extend within the nasopharynx and/or extend to the base of the skull, palate, nasal cavity or oropharynx and distant metastases can arise in bone, lung, mediastinum and liver [1,2]. Sinonasal carcinoma could arise from various tissues within the nasal cavity, including lymphoreticular, epithelial and nonepithelial tumors [3]. Sinonasal Papillomas are a rare disease which have an annual incidence of 0.6/105 in defined geographical regions. The investigations of these tumors have shaded the light on a viral agent as an etiological candidate, such as EBV [4]. In addition, an outdoor and industrial occupations are the major risk factors for sinonasal Papillomas development [5]. Inflammatory Polyps lesions are affecting 1-4% of all population [6]. Where they arise as an outgrowth from the mucosal membrane of nasal cavity and paranasal sinuses as a result of an allergic or chronic inflammation [7].
Epstein Barr virus (EBV) also has been shown to be associated to many human cancers, such as nasopharyngeal carcinoma, burkitt’s lymphoma and Non-Hodgkin lymphoma etc. [8,9]. The Epstein Barr virus Early Repeats (EBER1 and EBER2), have 167 and 172 nucleotides long in consequence with 54% sequence homology between them. EBERs were used for the recognition of EBV infected cells in tissues by CISH and are a reliable marker indicating the presence of EBV. EBER-1 interacts with interferoninducible protein kinase R (PKR), inhibiting its activation and protecting infected cells from IFN induced apoptosis and induce the expression of growth factors that promote cell survival [10,11].
Human cytomegalovirus (HCMV) has an implications in the etiology of several human cancers; including cervical carcinoma, prostatic adenocarcinomas and colonic and brain cancers [12]. HCMV infection might have role in building up the tumor cells through protection of certain tumor cells from apoptosis and modulating angiogenesis [13]. The phosphoprotein 65 (Pp65) was used as a reliable marker indicating the presence of HCMV and the recognition of HCMV infected cells in tissues by CISH technique.
Materials and Methods
This study enrolled a total number of one hundred fiftythree (153), formalin-fixed, paraffin-embedded blocks from nasopharyngeal and sinonasal tissues. They were collected from archives of histopathological laboratories of several hospitals in Baghdad. Some of these tissues block were related to the past few years (2015, 2016, 2017 and 2018) and are including 18 sinonasal carcinoma, 65 nasopharyngeal carcinomas, 35 sinonasal inverted Papillomas and 35 nasal inflammatory polyps in addition to matched 30 healthy sinonasal tissue blocks obtained from patients subjected to nasal bridge reconstruction operation as a control group.
The diagnoses were based on accompanied pathological reports of the corresponding patients. The age of patients and controls (120 males, 63 female) ranged between (17-83) years. Histopathological sections were made for biopsies and stained by hematoxylin and eosin for final definitive diagnosis. The detection of EBV and HCMV by Chromogenic in situ hybridization (CISH) reactions kit (purchased from Zyto Vision GmbH. Fischkai, Bremerhaven. Germany) (Cat. Numbers: T-1061-40) was done on 5μm tissue sections by using digoxigenin-labeled oligo-nucleotides probe which targets EBV-EBERs-RNA and HCMV-pp65 DNA (Cat. Number: T-1114-400 and T-1113-400) (Zyto Visions/Germany).
The detailed methods for performing CISH reactions were conducted according to the instructions of the above manufacturing company. The CISH signal patterns were classified as punctuate, diffused and mixed. The punctuate pattern, is, a dot-like signals or tiny particles that are scattered in the nucleus, this pattern refers to the integrated viral genome, and the diffused pattern, appears as a large globular with homogeneous strong staining in the nucleus or cytoplasm, this pattern indicates an episomal viral genome, while the mixed patterns represent the integrated and episomal viral genomes which appear together in the same sample. The quantification of CISH signal patterns (punctuated and diffused) were assessed under light microscopy at (100X) to visualize where virus-associated dsDNA localized in cytoplasm or nucleus by screening ten field in tissue loaded on chargeable slide by using the mean percentage of viral location [14]. Scoring and intensity of signals were done according to [8].
Statistical analysis
In this study, SPSS program (version-21) was used for statistical analysis where ANOVA test and LSD were used to assess the significances between variables.
Results
Distribution of EBER-CISH Signals patterns among sinonasal and nasopharyngeal tissues
A-Punctuated pattern: As shown in (Table 1) and (Figure 1,2) of the punctuated patterns in the sinonasal and nasopharyngeal cancer tissues, the mean of punctuators among tissues with nasopharyngeal carcinoma was higher (20.43) than the mean of punctuators in sinonasal carcinoma group (17.00), and the mean of sinonasal papilloma and inflammatory nasal polyp groups (13.29) and (6.51), respectively. In comparison, the mean of punctuators pattern in healthy control tissues was (2.30). Accordingly, there were highly significant statistical differences (P<0.01) between different groups.
Table 1: Distribution of punctuated and diffused patterns of EBER-CISH patterns among Studied groups.
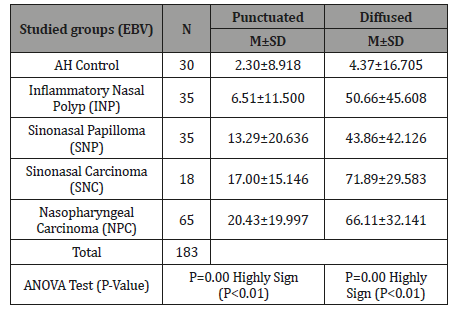
Table 2: LSD Statistical Analysis of Punctuated EBER-CISH Signals.
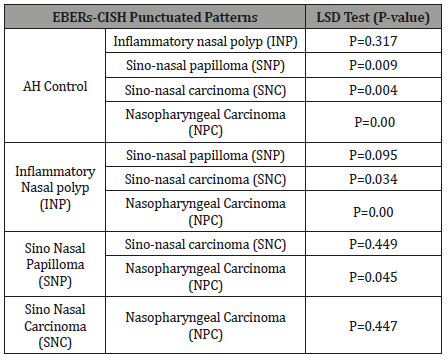
As detailed in (Table 2) a highly significant statistical differences were noticed between SNP, NPC, SNC and healthy control group and between NPC and INP groups. On the other hand, there were significant differences between SNC and INP and between NPC and SNP groups.
B-Diffused pattern: (Table 1) and (Figure 1,2) showed a diffused pattern in sinonasal and nasopharyngeal cancer groups, the mean of diffused form in malignant sinonasal carcinoma tissues was higher (71.89) than the mean in nasopharyngeal carcinoma tissues (66.11) and both were higher than the mean in the benign tumors in sinonasal papilloma and inflammatory nasal polyp groups (43.86 and 50.66), respectively. Whereas, the mean of diffused pattern in healthy control tissues was (4.37). Statistically there were a highly significant differences (P<0.01) between different groups.

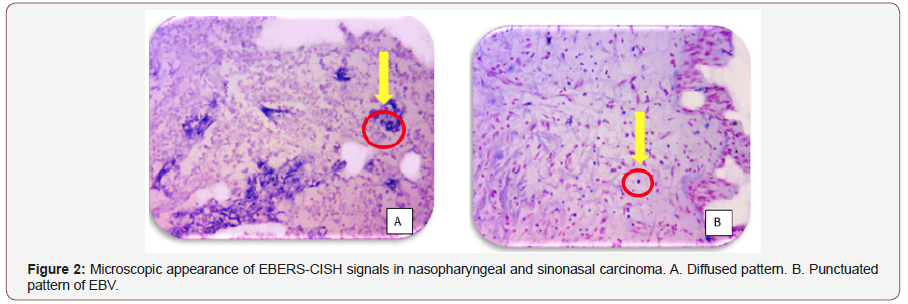
Table 3: LSD Statistical Analysis of Diffused Patterns of EBER-CISH Signals.
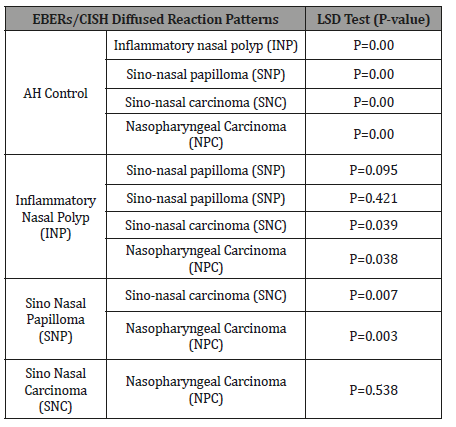
In detail there were a highly significant differences between SNP, INP, NPC, SNC and healthy control group and highly significant differences between each of (NPC and SNC) when compared to SNP group. Furthermore, significant differences between each of (NPC and SNC) and INP group. While there were no significant differences neither between SNC and NPC nor between SNP and INP groups (Table 3).
Patterns of HCMV-DNA distribution among studied sinonasal and nasopharyngeal tissues
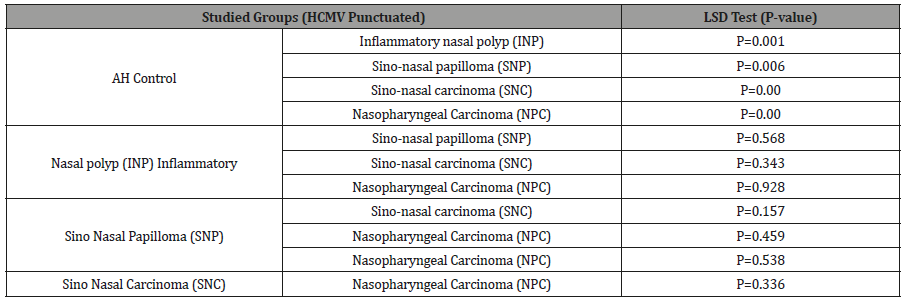
Punctuated Pattern: (Table 4) and (Figure 3,4) show that the mean of punctuated pattern in malignant sinonasal carcinoma tissues was higher (21.56) than the means (16.94) of this pattern in nasopharyngeal carcinoma tissues and each of sinonasal papilloma and inflammatory nasal polyps (14.14 and 16.60) respectively, in comparison it was (1.80) in healthy control tissues. Statistically, highly significant differences (P<0.01) were recorded between different groups. Highly significant differences were recorded between the groups of SNPs, INP, NPC, SNC and the healthy control group, while there were no significant differences between all other groups and as it shown in (Table 5).
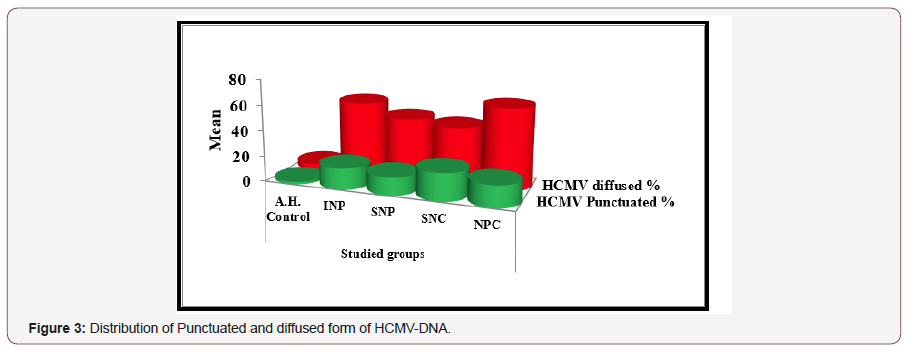
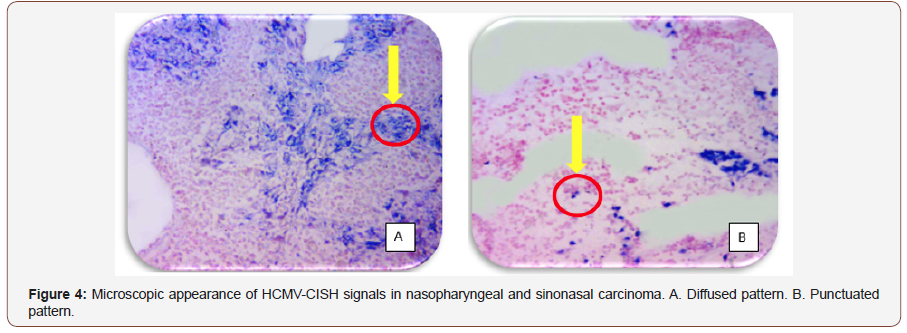
Table 4: Distribution of Punctuated and Diffused Pattern of HCMVpp65-DNA among the Studied Tissues.
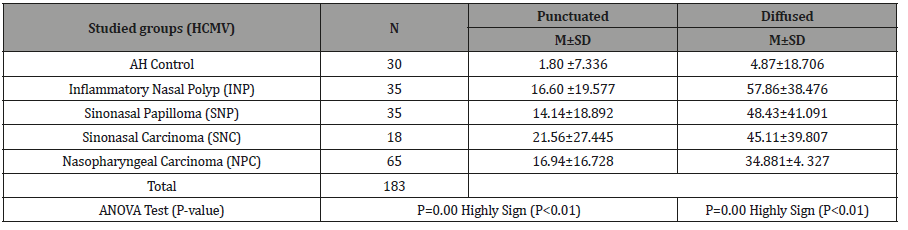
Table 5: LSD Statistical Analysis of Diffused Patterns of EBER-CISH Signals.
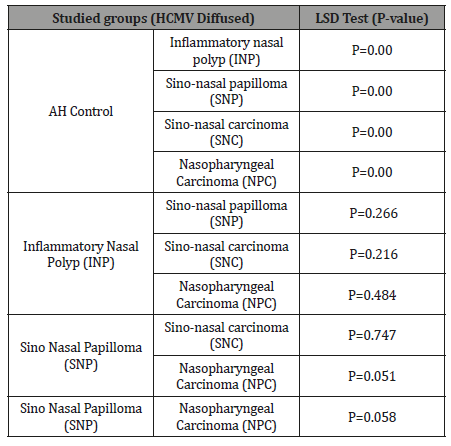
B-Diffused Pattern:The mean of diffused pattern in tissues with malignant sinonasal carcinoma was higher (45.11) than its mean in nasopharyngeal carcinoma tissues (34.881) and the mean in benign sinonasal papilloma (48.43), inflammatory nasal polyp (57.86), and the mean of this pattern in healthy control tissues (4.87). There were a highly significant differences (P<0.01) among these groups (Table 4) (Figure 3,4). These differences were highly significant between SNP, INP, NPC, SNC and healthy control groups, while there were no significant differences between the rest of groups (Table 6).
Table 6: LSD Statistical Analysis of Diffused Pattern of HCMV pp65- DNA-CISH Signals.
Discussion
Epstein Barr virus integration into host genome has been reported by [15-17]. Chromosomal integration of EBV genome represent a state of interaction between virus and host cells, this may reflect chromosomal instability that induce further genetic change in tumor cells and cause frequent DNA recombination which promote EBV integration into fragile DNA sites, that effect cell cycle genes such as tumor suppressor genes and cause accumulation of mutations that leads to transformational changes and induce oncogenesis as reported by [18]. Sugden and Dheekollu et al. [19,20] reported episomal EBV genome in NPC and some other tumors. The EBV episomal replicate independently of host cell genome and transfer to generations and in some instances, it recombines with the human genome to create one or more chromosomal integrations of varying structure with respect to viral and host breakpoints and cause accumulation of mutations and induce toumergenesis [21].
In regard to HCMV, both patterns were found in all of the examined tissue specimens, however the percentages of diffused pattern subsets of NP and SN tissues were higher than the subsets of these tissues which showed punctuated patterns. These findings indicate that this virus has established a persistent/latent state in all these benign and malignant tissues. In addition, this nuclear localization of such punctuated pattern of CISH found in the present study is an indication of viral genome integration in the cellular DNA, which is regarded as an irreversible as well as a critical step or process that associated in the transformation of human cellular DNA inevitably [22-24].
In a previous study by [25,26] also detected HCMV nucleic acids predominantly in the nuclear localization of the cancerous tissues. Moreover, nuclear integration HCMV-DNA of tumor tissues was regarded as an early event in the mutagenic process of their involvement associated with viral promotion of DNA damage and/or tumor initiation [27,28]. Furthermore, viral integration and insertional mutagenesis was found to be involved in tumor progression via the dysregulation of cellular processes, an effect of viral proteins at later stages modulating signaling pathways are involved in cell proliferation, inhibiting apoptosis, increasing rates of immortalization [29,30].
However, episomal genomic form observed in this study could have the chance to replicate as well as have the chance of integration, (although to a lesser extent) causing genomic instability and tumerigenesis [31,32]. One can concludes that these tissues were under the potential effects of being transformed, (in association with many other risk factors in the multistep development of such SN and NP carcinogenesis). Since it was revealed that the virus has either initiatory, co-factor or a promoter role in viral carcinogenesis [22].
Acknowledgements
None.
Conflict of interest
No conflict of interest.
References
- Wang R, Tan Y, Wang X, Ma L, Wang D, et al. (2014) Prognoses and long-term outcomes of nasopharyngeal carcinoma in Han and Uyghur patients treated with intensity-modulated radiotherapy in the Xinjiang Autonomous Region of China. PloS one 9(11): e111145.
- Al Shemmary AM (2015) Molecular Study of Epstein Barr Virus, P16 and Bcl2 Gene Expression in Tissues from Nasopharyngeal Carcinoma Patients, MSC thesis, University of Babylon, Iraq.
- Jain S, Jadav K (2017) Histopathology of soft tissue tumors in association with immunohistochemistry. International Journal of Biomedical and Advance Research 8(08): 327-336.
- Wang MJ, Noel JE (2016) Etiology of sinonasal inverted papilloma: A narrative review. World journal of otorhinolaryngology head and neck surgery 3(1): 54-58.
- Lun Sham C, Lee DL, Van Hasselt CA, Tong MC (2010) A case-control study of the risk factors associated with sinonasal inverted papilloma. Am J Rhinol Allergy 24(1): 37-40.
- Koskinen A, Salo R, Huhtala H, Myller J, Rautiainen M, et al. (2016) Factors affecting revision rate of chronic rhinosinusitis. Laryngoscope investig otolaryngol 1(4): 96-105.
- Knör M, Tziridis K, Agaimy A, Zenk J, Wendler O (2015) Human papillomavirus (HPV) prevalence in nasal and antrochoanal polyps and association with clinical data. PloS one 10(10): e0141722.
- Li YQ, Khin NS, Chua ML (2018) The evolution of Epstein-Barr virus detection in nasopharyngeal carcinoma. Cancer biol Med 15(1): 1.-5
- Tsang CM, Tsao SW (2015) The role of Epstein-Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. Virol Sin 30(2): 107-121.
- Agnieszka S (2016) Genetic analysis of the role of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) in B cell transformation. PhD thesis, Imperial College London/Department of Medicine P (40).
- Fitzsimmons L Kelly GL (2017) EBV and apoptosis: the viral master regulator of cell fate? Viruses 9(11): 339.
- Ahmed HG, Suliman RSA, Ashankyty IM, Albieh ZA, Warille AA (2018) Role of human Cytomegalovirus in the etiology of nasopharyngeal carcinoma. J Cancer Res Ther 14(3): 583-586.
- Biolatti M, Dell'Oste V, Pautasso S (2018) Human cytomegalovirus tegument protein pp65 (pUL83) dampens type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J virol 92(6): e01774-17.
- Khalid H (2019) Spatiotemporal Dynamics of a Generalized Viral Infection Model with Distributed Delay and CTL Immune Response. Computation 7(21): 1-16.
- Gao J, Luo X, Tang K, Li X, Li G (2006) Epstein-Barr virus integrates frequently into chromosome 4q, 2q,1q and 7q of Burkitt's lymphoma cell line (Raji). J Virol Methods 136(12): 193-199.
- Yu HX, Liu G (2014) Malignant transformation of sinonasal inverted papilloma: a retrospective analysis of 32 cases. Oncol lett 8(6): 2637-2641.
- Xiao K, Yu Z, Li X, Tang K, Tu C, et al. (2016) Genome-wide analysis of Epstein-Barr virus (EBV) integration and strain in C666-1 and Raji cells. J Cancer 7(2): 214-224.
- Chang Y, Chang SS, Lee HH, Doong SL, Takad K, et al. (2004) Inhibition of the Epstein–Barr virus lytic cycle by Zta-targeted RNA interference. J gen virol 85(6): 1371-1379.
- Sugden B (2014) Epstein-Barr virus: the path from association to causality for a ubiquitous human pathogen. PLoS bio, 12(9): e1001939.
- Dheekollu J, Malecka K, Wiedmer A, Delecluse HJ, Chiang AK, et al. (2017) Carcinoma-risk variant of EBNA1 deregulates Epstein-Barr virus episomal latency. Oncotarget, 8(5): 7248-7254.
- Reisinger J, Rumpler S, Lion T, Ambros PF (2006) Visualization of episomal and integrated Epstein‐Barr virus DNA by fiber fluorescence in situ hybridization. International journal of cancer 118(7): 1603-1608.
- Aldrubi IF, Abdul Majeed BA, Mukhlis FA, Al-nasri H, Alsultan RA (2015) The validity of real time PCR and in situ hybridization in the Detection of JCV in colonic Biopses of patients with colorectal cancer. International Microbiology Application Science 4(12): 107-120.
- Mietzner TA, Carroll KC, Hobden JA, Miller S, Morse SA, et al. (2016) Jawetz, Melnick & Adelberg's Medical Microbiology (27th), Chapter 29, Mc Graw Hill Education, Pp: 618-635.
- Volpi CC, Gualeni AV, Pietrantonio F, Vaccher E, Carbone A, et al. (2018) Bright field in situ hybridization detects gene alterations and viral infections useful for personalized management of cancer patients. Expert Rev Mol Diagn 18(3): 259-277.
- Chan BW, Woo JKS, Liew CT (2002) Cytomegalovirus infection of the nasopharynx. J Clin Pathol 55(12): 970-972.
- Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, et al. (2010) Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae 1(1): 8-15.
- Richardson AK, Currie MJ, Robinson BA, Morrin H, Phung Y, et al. (2015) Cytomegalovirus and Epstein-Barr virus in breast cancer. PloS one 10(2): e0118989.
- Karamitros T, Van Wilgenburg B, Wills M, Klenerman P, Magiorkinis G (2018) Nanopore sequencing and full genome de novo assembly of human cytomegalovirus TB40/E reveals clonal diversity and structural variations. BMC genomics 19(1): 577-583.
- Herbein G (2018) The human cytomegalovirus, from on comodulation to oncogenesis. Viruses 10(8): 408-413.
- Litjens NH, Van Der Wagen L, Kuball J, Kwekkeboom J (2018) Potential Beneficial effects of Cytomegalovirus infection after Transplantation. Front Immunol 1(9): 389-398.
- Weisbach H, Schablowsky C, Vetter B, Gruska I, Hagemeier C, et al. (2017) Synthetic lethal mutations in the cyclin A interface of human cytomegalovirus. PLoS pathog 13(1): e1006193.
- Ravichandran S, Kim YE, Bansal V, Ghosh A, Hur J, et al. (2018) Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS pathog 14(9): e1007334.
-
Majid Mohammed Mahmood, Marwa Mohammed Ali Jassim, Saad Hasan Mohammed Ali. Localization of Herpetic Viruses Patterns in Sinonasal and Nasopharyngeal Malignant and Benign Tumors. On J Otolaryngol & Rhinol. 1(5): 2019. OJOR.MS.ID.000524.
-
Oncomodulatory, Sinonasal, Nasophary, Tumor, Nasal cavity, Paranasal sinus, Sinonasal papilloma, DNA sites, Human genome, Cellular DNA
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.