 Research Article
Research Article
Isoprenoid Quinone Composition of the Genus Microbacterium and Related Strains
Mohammed Nizar Battikhi*
De Montfort University, School of life Science, Leicester University Department of Microbiology, England
Moh’d Nizar Battikhi, 1017-1645 De Maisonneuve O, Montreal H3H 2N3, QC, Canada.
Received Date: June 12, 2019; Published Date: July 12, 2019
Abstract
The aim of this study is to compare pattern of Isoprenoid quinine of unspecified bacterial isolates from dairy product to reference strains type species of the genus Microbacterium.
Method: Twenty-eight strains of coryneform and related bacteria were studied for isoprenoid quinine content. Extraction of 100 mg of freezedried cells by chloroform: methanol (2:1v/v), extract was then run onto analytical TLC plates coated with o.5 mm Merck Silica Gel HF245.
Results: Out of the twenty-eight reference and unspecified strains M. lacticum and M. flavum are quite distinct and that the predominant of MK-8 in BL77/18. BL77/19 and BL77/20 support that these strains are related to M. flavum.
Keywords: Micro bacterium; Isoprenoid quinone; TLC
Introduction
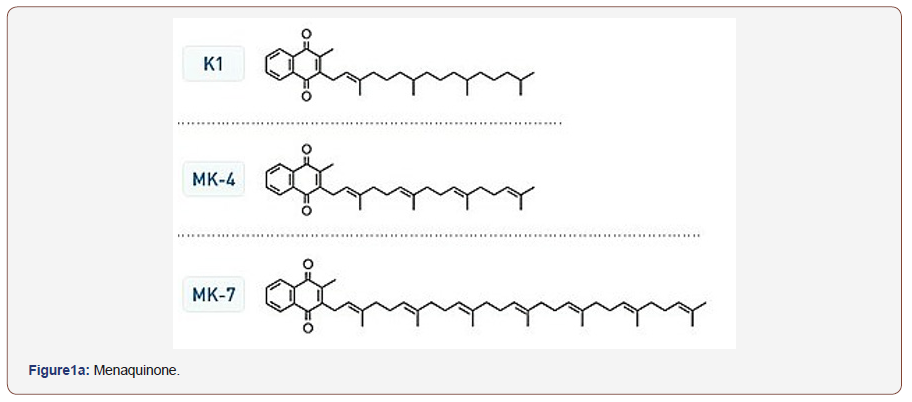
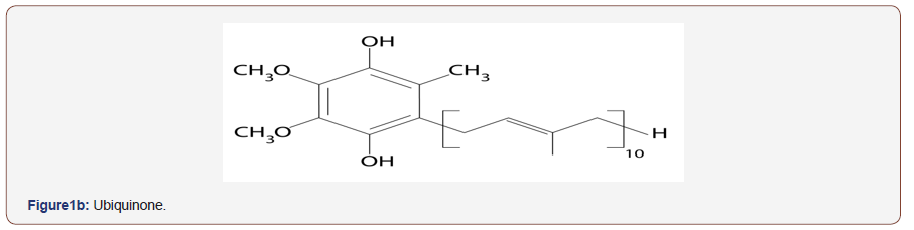
Coryneform bacteria classification in Bergy’s Manual of Bacteriology [1] accommodate different genera as Cellumonas, Arthrobacter, Corynebacterium and Microbacterium described as incertea sedis. Other motile plant pathogens were added to coryneform group [2,3]. Numerical phonetic studies improved classification of coryneform [4,5]. Other markers were used to improve taxonomic position of coryneform bacteria such as DNA base composition determination [4,6-10], wall analysis [11-15] and mycolic acid analysis [2,15,16]. The use of additional marker (isoprenoid quinone) was recommended by [16,17] to improve classification of coryneform and related taxa. The analysis of the isoprenoid quinone of coryneform bacteria has been a useful aid to establish the taxonomic relatedness of bacterial strains [2, 3,17-20]. These quinines can be divided into menaquinones and ubiquinones abbreviation of their data is given in (Figure1a). Substantial investigation amongst coryneform have been carried out on the distribution of quinines bearing isoprenoid side chain. Bacterial quinones of this type can be sub-divided into menaquinones (2 methyl-3-polyisoprenyl 1-1,44 naphthoquinones and ubiquinones (co-enzyme Q; 12,3-dimethoxy-5-methyl-1-6- polyprenyl-1-1,4 benzoquinone (Figure1b). The menaquinones (related to vitamin K) show structural variation at C-2 and ,more especially, at C-3.Variation at C-3 include the length and degree of un-saturation of the polyprenyl side-chain [22] and the presence of hydroxyl [21] and epoxide groups [22]. Similarly, ubiquinones show variation principally in the structure of polyprenyl sidechain [23]. Therefore, the structure of these compound might be of great value in classification of coryneform and related bacteria [23]. Other studies demonstrated that organisms can be clustered based on the isoprenoid side chain of the menaquinones [2,3,20,24,25]. From their analysis Minnikin et al [22] concluded that MK_8(H2) was the major menaquinones of the animal associate corynebacterium. Microbacterium flavum, with consistently shows affinities with the animal associated corynebacteria in numerical phonetic studies, also contains MK-8(H2). They also reported that Micro bacterium ammoniaphilum contained MK-9(H2). Isoprenoid quinines are membrane-bound compounds (lipid molecules) found in nearly all living organisms. It marked structural variation depending upon the microbial taxon [27-30]. Therefore, organisms can be clustered based on isoprenoid side chain of their menaquinones. and hence differentiation between various taxa could be improved [3,17,18,27].
Method
Twenty-eight strains of coryneform and related bacteria were collected for the study. These were obtained from public and private culture collections (Table 1). Each strain was given number for ease handling. Several strains representing different taxa defined in study [4,5] where numerical phonetic surveys of coryneform and related bacteria were included as references. The unidentified strains were received from the National Institute of Research in Dairying, Sheffield, Reading and food Research institute, Norwich. All strains were originally obtained in freeze dried cultures.
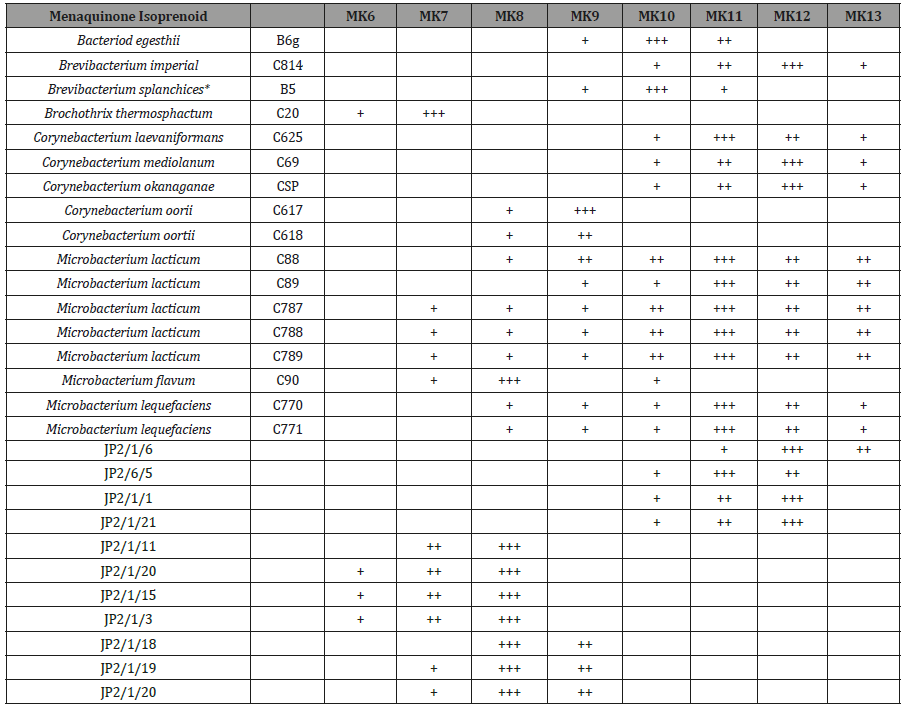
Table 1: Menaquinone composition of strains.
General Growth Media
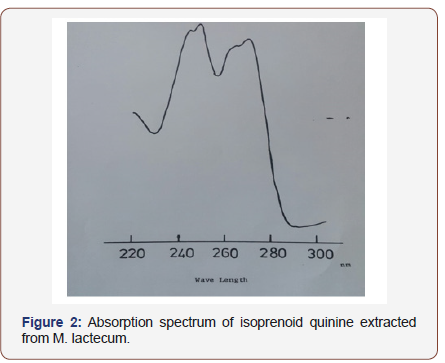
Strains were routinely subculture at three weeks intervals onto nutrient agar (Difco) plates and stored at 4C. After 2-3 days incubation at 37 °C. For broth culture Nutrient broth Media (Oxoid) media was and culture were stored at lyophilized aliquots in glass vials for future used were they resuscitated by transfer small amount of nutrient onto nutrient agar plates and incubated for 7 days. Nutrient agar (Difco) and nutrient broth (Oxoid) were used for growing all isolates. All media were sterilized by autoclaving at 121 °C for 15min. Culture were incubated for 24-48 H at 37 °C. Cultures were harvested by centrifugation at 8,000 rpm for 20min, in an MSE high speed M-18 centrifuge, washed in distilled water and re-harvested Organism were lyophilized and stored anhydrously as a fine powder until required and freeze dried for future used. Isoprenoid quinone analysis done by extraction of 100mg potions freeze dried organisms by a method described by Collins et al. [17] that proved to be satisfactory for the analysis of lipids. At the preparation phase ,components with chromatograph with vitamin K were the only isoprenoid quinines which were detected. Ultraviolet spectroscopy of the eluted quinines showed absorption maxima at 242,248,260,270 and 326nm (Figure 2), feature which are in accordance with the published data for menaquinones [28] where 100mg of freeze dried cells were mixed with 20ml chloroform: methanol (2:1v/v) in a 50ml conical flask and the suspension stirred for16-18h in the dark (isoprenoid quinine are susceptible to strong light). Organisms were removed by filtration and the extracts were evaporated to dryness under reduced pressure and teem below 37 °C. Components were detected by observing chromatograms under short-wave (254nm) ultra-violet light. The dried extracts were prepared as described in above then were re-suspended in small volume of chloroform: ethanol (2:1 v/v) and spotted onto analytical TLC plates coated with 0.5mm Merck Silica Gel HF245 (prepared by coating plates with slurry contains 40g gel HF245 per 100ml distilled water and drying overnight at 65 °C).
Plates were developed in petroleum ether (BP 60-80): diethyl ether (85:15 v/v). In this solvent system, the menaquinones migrate further (Rf 0.7) than ubiquinones (Rf 0.4). Vitamin K (sigma) an Ubiquinone’s -50 (BDH) were included as standard. The isoprenoid quinines were also isolated by eluting with chloroform from preparative TLC plates (1 mm, Merck silica Gel HF245) using the same solvent system. The quinines were detected by brief irradiation with ultraviolet light (245nm) when they appeared as dark brown spots on a bright green fluorescent background. Eluted samples were evaporated to dryness by a method based on Collins et al [17] proved to be satisfactory for analysis of lipids at the preparation phase, components with chromatographed with vitamin K were the only isoprenoid quinines which were detected. Ultraviolet spectroscopy of the eluted quinones showed maxima at 242, 248, 269, 270 and 326nm (Figure 2) feature which are in accordance with the published data from menaquinones [28]. The isoprenoid quinine was separated by reverse phase partition chromatography using appropriate standards. Components were detected by observing chromatogram under ultraviolet light (254nm).
Result
In describing the menaquinones of strains, the following nomenclature was adopted. The symbol MK denoting menaquinone is followed by a number (e.g. 8, 9 or 10) denoting the number of isoprene units attached as a side chain (Table 2). A wide range of manaquinones with varying numbers of isoprene units were characterized within the group of the menaquinones although only one type predominated of Corynebacterium, oortii (strain C617 and C618) was MK9, a result in agreement with other study [20]). Likewise, the major quinine of Microbacterium flavum (C90) was MK8, again in agreement with published data [17,29]. In this study the menaquinones of 28 reference strains of Micro bacterium, Corynebacterium and unknown isolates were examined. The five strains of M. lacticum (C88,C,89,C787,C788.C789) tested were all contain eleven and twelve isoprene units as the predominant side chain of their menaquinone in contrast M. flavum (C90) was shown to contain MK-8 as the predominant menaquinones which is in agreement with other study by [2]. These observations agree with conclusion that M. lacticum and M. flavum are quite distinct, likewise the major menaquinone (MK-7) of Brochothrix thermosphacta (Microbacterium thermosphactum) support the other evidence that this species is quite distinct from both M. lacticum and M. flavum. The presence of MK-11 and MK12 in Corynebacterium laevaniformans (C626) agrees with other study [20]. Strains JP2/1/6 and strain JP2/1/5 were both contain MK-11 and MK12, which is similar to M. lacticum that showed resemblances. M. liquefaciens (C770, C771), Corynebacterium mediolanum (C69) and Brevibacterium imperial (C25) all shown to contain MK-11 and MK-12 a feature consistent with earlier study [19]. The unnamed strains used in this study showed that the predominant of MK-8 in BL77/18. BL77/19 and BL77/20 support that these strains are related to M. flavum. The similar menaquinones profiles of strains JP2/1/1 and`JP2/1/21which contain MK-11 and MK-12 supports their taxonomic relatedness but suggest that they may not closely related to M.flavum. The predominant of MK-8 in strains.
Table 2: Isoprenoid quinine of coryneform and related bacteria (Abstracted from Minnikin et al. [3]).
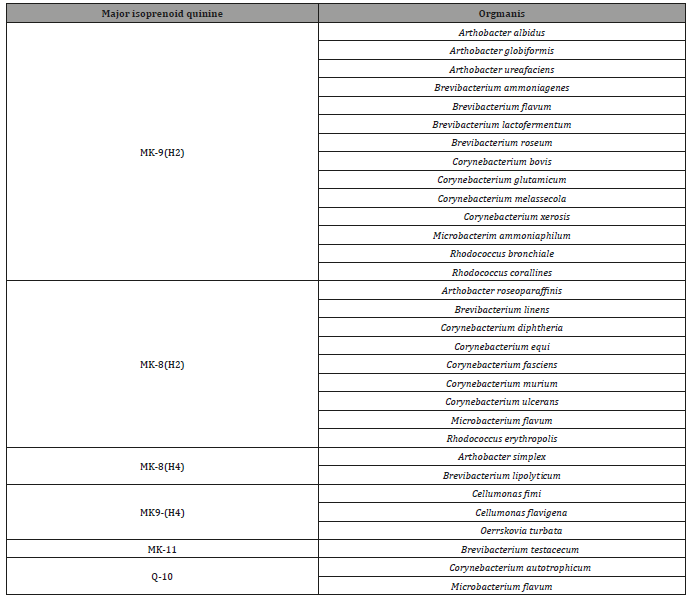
Discussion
The analysis of isoprenoid quinines of coryneform bacteria has been a useful aid to establishing the taxonomic relatedness of strains [2,18-21,26] These quinines can be divided into menaquinones and ubiquinones. By far the majority of coryneform contains menaquinones although different groups produce menaquinones with different numbers of isoprene units and may also differ in their degree of saturation. These differences appear to be taxonomically significant [26]. Thus, organism regarded as closely related on other evidence (numerical taxonomic data, cell-wall composition,) often have similar menaquinones while unrelated organisms frequently contain different components.
In this study the menaquinones of 28 strains were examined. The five strains of M. lacticum (C88, C89, C787, C788, C789) used all contained eleven and twelve isoprene units as the predominant side chain of their menaquinones. In contrast M. flavum (C90) was shown to contain MK-8 as the predominant menaquinone which agrees with other study [26]. These observations agree with the conclusion that M. lacticum and M. flavum are quite distinct. Likewise, the major menaquinone (ML-7) of Brochothrix thermosphata (Microbacterium thermosphactum) supports the other evidence that this species is quite distinct from both M. lacticum and M. flavum. The presence of MK-11 and MK-12 in Corynebacterium lave K-11 and MK-12 which showed their relatedness to each other and similar to M. lacticum. The significant of manaquinone in taxonomy was reported by Collins et al [18].
They showed that, amongst other criteria, the curtobacteria can be divided into two groups based on their menaquinones. The significant of menaquinone marker in taxonomy was clearly appeared by showing relatedness of unknown isolates as BL77/18, BL77/19. BL77/20 to each other and to M. flavum since they showed MK-8. Likewise, the similar menaquinones profiles of strain JP2/1/1 and JP2/1/21 which contain MK-12 supports their similar taxonomic position but they are closely related to M. flavum. Likewise, our result in this study showed the presence of MK-9 in strains JP2/1/11, JP2/1/20, JP2/1/15 and JP2/1/3 which suggest resemblance of these strains.
Conclusion
This study showed the significant of isoprenoid quinine as marker of clustering different strains according to their menaquinone content which may lay base line for future taxonomy. However, other markers may be need for further support.
Acknowledgement
None.
Conflict of Interest
No conflict of interest.
References
- Rogosa M, Cummins CS, Lelliot RA, Keddie RM (1974) Coryneform group of bacteria In: RE Buchana, NE Gibbson (Eds.), Bergy’s manual of determinative bacteriology (8th edn.), William, Wilkins, Baltimo.
- Yamada Y, Inouye C, Tabara Y, Kondo K (1976) The menaquinone system in the classification of coryneform and nocardioform bacteria and related organisms. Journal of General and applied Microbiology 22(4): 203-214.
- Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100(2): 221-230.
- Bousfield IJ (1972) A taxonomic study of some coryneform bacteria. J Gen Microbiol 71(3): 441-455.
- Skyring GW, Quadling C (1970) Soil bacteria: Principal component analysis and guanine-cytosine contents of some arthrobactercoryneform soil isolates and some named cultures. Can J Microbiol 16(2): 95-106.
- Yamada K, Komagata K (1970) Taxonomic studies on coryneform bacteria III. DNA base composition of coryneform bacteria. J Gen Appl Microbiol 16: 215-224.
- Bowle IS, Griger MR, Duncklay GG, Loutit MW, Loutit JS (1972) The DNA base composition and fatty acid constitution of some Gram-positive pleomorphic soil bacteria. Soil biology and Biochemistry 4(4): 397-412.
- Crombach WH (1972) DNA base composition of soil arthrobacter and other coryneform from cheese and sea fish. Antonie Van Leeuwenhoek 38(2): 105-120.
- Starr MP, Mandel M, Murata N (1975) The phytopathogenic coryneform bacteria in the light of DNA-base composition and DNA-DNA segmental homology. J Gen App Microbiol 21(1): 13-26.
- Schleifer KH, Kander O (1972) Peptdoglycan types of bacterial cell wall and their taxonomic implications. Bacteriol Rev 36(4): 407-477.
- keddie RM, leask BGS, Grainger JM (1966) Comparison of coryneform bacteria from soil and herbage. Cell wall composition and nutrition. Journal of Applied Bacteriology 29 (1): 17-43.
- Yamada K, Komagate K (1972a) Taxonomic studies of coryneform bacteria IV Morphological, cultural, biochemical and physiological characteristics. J Gen Appl Microbiol 18: 399-416.
- Yamada K, Komagata K (1972b) Taxonomic studies one form of coryneform bacteria V Classification of Coryneform bacteria. J Gen App Microbiol 18: 417-431.
- Keddi RM, Cure GL (1977) The cell wall composition and distribution of free mycolic acids in named strains of coryneform bacteria and in isolates from various natural sources. J Appl Bacteriol 42 (2): 229-252.
- Goodfellow M, Collins MD, Minnikin DE (1976) Thin layer chromatographic analysis of mycolic acid and other long-chain components in whole organism methanolysates of coryneform and related taxa. J Gen Microbiol 96(2): 351-358.
- Collins Thompson DI, Witter LD, Sorhuge T, Ordal ZJ (1972) Taxonomic consideration of Micro bacterium lacticum, Microbacterium flavum and Microbacterium thermosphactum. International Journal of Systematic Bacteriology 22 (2): 65-72.
- Collins MD Lipods in Corynebacterium taxonomy PhD. Thesis. Newcastle upon Tyne, UK.
- Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing paptidoglycan based on 2,4 diaminobutyric acid. J of Applied Bacteriology 48(3): 459-470.
- Minnikin DE, Goodcfellow M, Collins MD (1978) Lipids composition in the classification and identification of nocardia and related taxa. In: M Brownell GH, Serrato JA (eds.), The biliology of Nocardia Goodfellow, New York and London Academic press, pp: 160-69.
- Bousfield IJ, Callely (1996) AC Special publication of the society for General Microbiology I. Coryneform Bacteria London Academic Press.
- Thomson RH (1971) Naturally occurring quinine. New York and London Academic press.
- Allines CF, Frake H, Hiryama O (1967) Identification of plastoquinone and two naphthoquinones in anacystis nidulans by NMR and Mass spectroscopy. Biochem Biophys Res Commun 26(5): 562-568.
- Allines CF, Frake H, Hiryama O (1967) Identification of plastoquinone and two naphthoquinones in anacystis nidulans by NMR and Mass spectroscopy. Biochem Biophys Res Commun 26(5): 562-568.
- Gale PH, Arison BH, Trenner NR, Fage AC, Fokers K (1963) Characterization of vitamin K9(H) from Mycobacterium phlei. Biochemistry 2: 200-203.
- Collis MD, Goodfellow M, Minnikin DE (1979) Isoprenoid quinine in classification of coryneform and related bacteria. J Gen Microbiol 110(1): 127-136.
- Collis MD, Goodfellow M, Minnikin DE (1979) Isoprenoid quinine in classification of coryneform and related bacteria. J Gen Microbiol 110(1): 127-136.
- Beatrycze Nowick, Jerzy Kuuk (2010) Occurrence, biosynthesis and function of isoprenoid quinines. Biochim Biophys Acta 1797(9): 1587- 16.
- Hiraishi A (1999) Isoprenoid quinine as biomarkers of microbial population in the environment. J Biosci Bioeng 88(5): 449-460.
- Collins M, Jones D (1982) Distribution of isoprenoid quinine structural types in bacteria and their taxonomic implication. Microbiol Rev 45(2): 316-354.
- Dunfy PJ, Brodie AF (1971) The structure and function of quinines in respiratory metabolism. Methods in enzymology 18 (c): 407-461.
-
Mohammed Nizar Battikhi. Isoprenoid Quinone Composition of the Genus Microbacterium and Related Strains. Arch Clin Case Stud. 1(4): 2019. ACCS.MS.ID.000518.
-
Micro bacterium, Isoprenoid quinone, TLC, Cellumonas, Arthrobacter, Corynebacterium, Polyprenyl, Bacteria, Strains, Organisms, Coryneform.
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






