 Research Article
Research Article
DNA Interaction Study of Some Symmetrical 1,2-Phenylenediamine Schiff’s Base Derivatives as New Potential DNA Intercalators Using Ethidium Bromide Competition Fluorescent Assay
Sofian S Mohamed1, Inass A Sadawe1, Nisreen H Meiqal1, Abdulathim A Alshoushan2, Aisha S Aboud1, Safa A Aboulqasim1, Aisha A Issa1, Salah M Bensaber1, Anton Hermann3 and Abdul M Gbaj1*
1Department of Medicinal Chemistry, Faculty of Pharmacy, University of Tripoli, Libya
2Department Food and Drug Control Centre (LFDA), Tripoli, Libya
3Department of Biosciences, University of Salzburg, Salzburg, Austria
Abdul M Gbaj, Associate Professor of Genetics and Biochemistry, Department of Medicinal Chemistry, Faculty of Pharmacy, Libya.
Received Date:July 12, 2019; Published Date:July 18, 2019
Abstract
A series of novel potential DNA intercalators of symmetrical 1,2-phenylenediamine Schiff’s base derivatives were synthesized. The binding properties of these compounds to genomic DNA (G-DNA) have been investigated by fluorescence spectroscopy. Ethidium binding to DNA significantly enhanced its fluorescence making it a convenient probe to evaluate DNA binding of many drugs. The addition of a DNA binding agent induces a progressive decrease in fluorescence of ethidium due to its displacement from the duplex. This also allows distinguishing non-intercalative binding agents from intercalating agents: agents having a huge site size groove binder need correspondingly smaller concentrations to saturate the sites. The results indicate that all the targeted compounds can interact with G-DNA, and among them, SW7 (1,1’-((1E,1’E)-(1,2-phenylenebis(azanylylidene)) bis(methanylylidene))bis(hthalen-2-ol)), showed the highest key selection vector (KSV) value, suggest that compound SW7, binds most strongly to G-DNA
Keywords: Fluorescence; DNA intercalation; Key selection vector
Introduction
Several biological experiments proposed that DNA is one of the principal cellular targets for numerous anticancer agents. Mainly, in cancer cells, DNA could be selectively damaged, as a result of interactions with anticancer agents, consequently blocking of cell division leads to cell death [1,2]. The molecules that interact with DNA are generally bound to DNA through non-covalent bonds by three main mechanisms: groove binding, intercalation or static electronic interactions. Static electronic interactions could be related to molecules that bind to the negatively charged DNA double helix at external sites through non-specific interaction. The second mode of interaction is called groove binding in which the targeting molecules interact with DNA in base edges of minor or major grooves [3-6]. The third mode of interaction is intercalation that is closely related to the antitumor ability of several anticancer drugs [7-9].
Intercalators are ligands interacting with the DNA double helix in a reversible manner. Many of them are currently used as powerful drugs for the treatment of breast and ovarian cancers, acute leukemia, and many others are still in clinical trials. Intercalating agents have several universal structural features, the most important is a planar polyaromatic system that inserts between DNA base-pairs with a marked preference for the purine-3’-pyrimidine-5’ sequence [10- 12]. Ethidium bromide is frequently utilized as a fluorescent tag (nucleic acid stain) in molecular biology for many procedures such as agarose gel electrophoresis. It intercalates with double-stranded DNA and leads to the deformation of the DNA which could affect biological processes, such as DNA transcription and replication [13,14].
Schiff’s bases (imines) are compounds that contain azomethine groups [-HC=N-] in their structure. They are formedby condensation of a dynamic carbonyl compound with a primary amine [1516]. Schiff’s bases show a large range of biological activities including antiviral, antibacterial, anti-inflammatory, antiproliferative, antifungal, antimalarial and antipyretic pharmacological activities [17-21]. We have recently reported on using microwave assisted synthesis and antimicrobial evaluation of symmetrical 1,2-Phenylenediamine Schiff’s base derivatives finding that some of the synthesized compounds showed antibacterial and antifungal activity [22]. Now our aim is to explore how different structural features of 1,2-Phenylenediamine Schiff’s base derivatives can affect the DNA binding capability using molecular modeling and key selection vector studies which may serve as a basis for understanding the molecular mechanism of action of the 1,2-Phenylenediamine Schiff’s base derivatives and can help to design of new chemotherapeutic molecules
Material and Methods
Chemistry
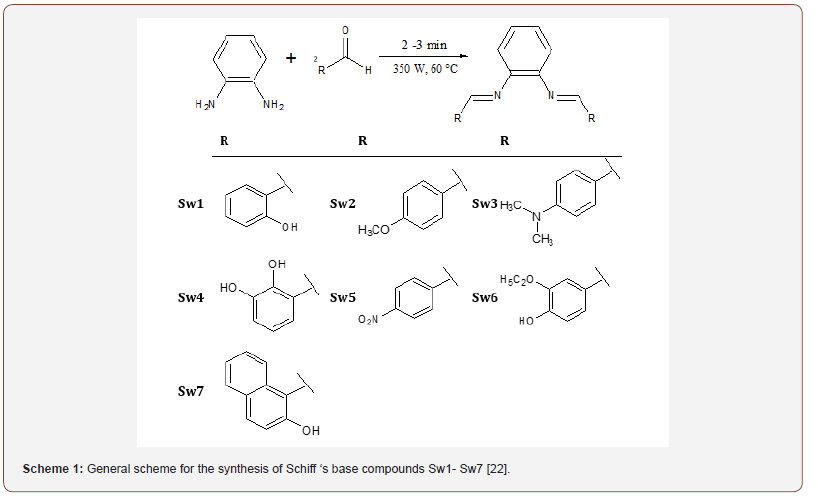
General procedure for synthesis of schiff ’s bases (Sw1- Sw7): Schiff’s bases were prepared according to our previously reported procedure by reacting of one mole of phenylenediamine and two moles of substituted aromatic aldehydes [22]. All reactants were mixed together and small amount of ethanol (5-6 ml) was added. This mixture was subjected to microwave irradiation at 350 watt for 2-3 minutes at a maximum of 60°C. After the reaction was completed (2-3 minutes), the mixture was left for cooling and the solid product (crude) was gathered by filtration and washed several times (5-6 washes) with ethanol and then vacuum dried. The gained product was recrystallized in ethanol and after drying that gave the clean pure product (Scheme 1). The crystalline product obtained were characterized as described previously by FTIR and 1H NMR [22] (Scheme 1).
DNA Binding Properties
To study how competently our synthesized compounds, interact with G-DNA, the compounds were investigated for their DNA binding ability using fluorescence emission spectroscopy. All experiments were conducted in Tris buffer (0.01M Tris, 0.1M NaCl, at pH 7.4), glass-distilled deionized water and analytical grade reagents were used throughout experiments. The pH values of all solutions were measured with a calibrated Jenway pH-meter model 3510 (Staffordshire, UK). All buffer solutions were filtered through Millipore filters (Millipore, UK) of 0.45 mm pore diameter.
Absorbance spectra: Absorbance spectra were measured on a Jenway UV-visible spectrophotometer, model 6505 (London, UK) using quartz cells of 1.00 cm path length. The UV-Vis absorbance spectra were recorded in the 200-500 nm range and a spectral bandwidth of 3.0 nm. For the final spectrum of each solution analyzed baseline subtraction of the buffer solution was performed. Genomic DNA was used in a concentration of 75 μg/ml. DNA was extracted from peripheral lymphocytes of anticoagulated blood (EDTA) samples by Proteinase K digestion and phenol/chloroform extraction [23]. The purity was determined by measuring the absorbance at 260/280nm indicating that the sample is free from protein contamination [23]. The concentration was assayed spectrophotometrically using 6600M-1cm-1 as a molar extinction coefficient at 260 nm
Fluorescence spectra and DNA-binding studies: Fluorescence emission and excitation spectra were measured using a Jasco FP-6200 spectrofluorometer (Tokyo, Japan) using fluorescence 4-sided quartz cuvettes of 1.00 cm path length. The automatic shutter-on function was used to minimize photo bleaching of the sample. The selected excitation wavelength for ethidium bromide was 480 nm. The emission spectrum was corrected for background fluorescence of the buffer. The ethidium bromide (EB) fluorescence displacement experiment wasperformed by sequential addition of aliquots of 1790μl Tris buffer, 10μl EB (final concentration of 72 μM), 100μl G-DNA from stock solutions (1.5mg/ml) and finally 10μl of compounds (SW1 to SW7, final concentration of 30 μM). Emission spectra were recorded for each system using excitation wavelengths of maximum fluorescence intensity determined for the systems to be 480 nm using a slit width of 5nm to examine alterations in emission spectra resulting from the complex construction of both systems. On construction of the full systems, the system was allowed to equilibrate for 30 minutes at room a temperature and emission spectra (500-730 nm) were recorded to monitor changes in EB intensity.
Prediction of ADMET properties and drug-likeliness: To verify drug-likeliness, canonical SMILES format of the synthesized molecules SW compounds were used in swiss ADME (http://www. swissadme.ch/index.php) [24]. The main bioavailability parameters were molecular weight (g/mol), lipophilicity (XlogP3), solubility (log S), polarity (Topological Polar Surface Area - TPSA in Å2), saturation (fraction of carbon atoms in Sp3 hybridization–Csp3) and flexibility (No of rotatable bonds). Absorption parameters such as Human Intestinal Absorption, Blood Brain Barrier, P-glycoprotein interaction (substrate or inhibitor) and metabolism (inhibitor or substrate) of the bio-active molecules with different cytochrome P450 enzymes were also estimated using admetSAR, which is based on QSAR data for prediction of absorption, distribution, metabolism, excretion and toxicity (ADMET) [25,26]. Drug-like molecules considering Ghose, Veber, Egan, Muegge and Lipinski rules and good ADMET properties were chosen as ligands in a consequent molecular docking procedure.
Molecular docking: The starting geometry of the synthesized SW compounds (SW1-SW7) was constructed using chem3D Ultra (version 8.0, Cambridge soft Com., USA). The optimized geometry of SW compound with the lowest energy was used for molecular docking. Crystal structures of the DNA fragment with the B-form 1bna, 102d, 1k2j and 1dne were downloaded from the Protein Data Bank (https://www.rcsb.org/structure/102D). Molecular dockings of SW compounds with B-DNAs were accomplished by Auto Dock 4.2 software from the Scripps Research Institute (TSRI) (http:// autodock.scripps.edu/). Firstly, polar hydrogen atoms were added into B-DNA molecules. Then, partial atomic charges of the B-DNA and SW molecules were calculated using Kollman methods [27]. In the process of molecular docking, the grid maps of dimensions (62Å X 62Å X 62Å) with a grid-point spacing of 0.376Å and the grid boxes is centered. The number of genetic algorithms runs and the number of evaluations were set to 100. All other parameters were default settings. Cluster analysis was performed on the results of docking by using a root mean square (RMS) tolerance of 2.0Å, which was dependent on the binding free energy. Lastly, the dominating configuration of the binding complex of SW compounds and B-DNA fragments with minimum energy of binding were determined.
Results and Discussion
Synthesis
The targeted compounds, symmetrical 1,2-phenylenediamine Schiff’s base derivatives, have been successfully synthesized using previously reported synthetic procedures [22]. Their chemical structures were confirmed by means of spectroscopic analysis as described previously [22].
ADMET properties
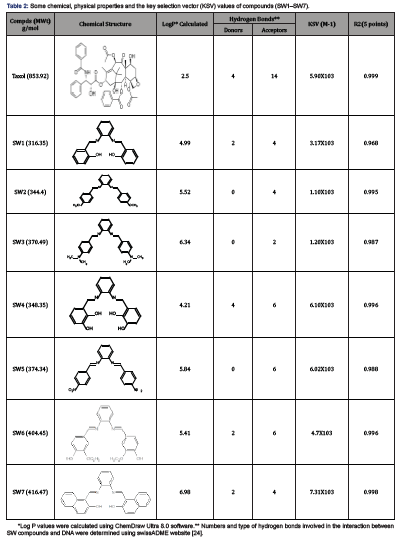
The pharmacokinetic properties, Lipinski’s rule of 5 and toxicity of all synthesized SW compounds were studied. Lipinski’s rule of five (L. Ro5) for drug-likeliness includes i) molecular mass should be below 500 kDa ii) log P value should be below 5 iii) hydrogen bond donorsites should be 5 or below iv) hydrogen bond acceptor sites should be 10 or below. An orally active compound should not have more than 2 violations of the Lipinski rule and have rotatable bonds (0-15) and polar surface area (7-200 Å). All the synthesized compounds reported in this paper were not found to violate the rule more than the maximum permissible limits and have rotatable bonds and polar surface area within the range thus proving their drug likeness properties. Lipophilicity (LogP o/w) values of all synthesized compounds were in range from 2.50 to 6.98 (Table 2), while their molecular weights are within the range 316.35-416.47 gram/mole. The compounds were found with optimum number of hydrogen bonding donors (0-4) and acceptors (2-6) as shown in Table 2. The achieved results of this part of our study indicate that most of the compounds follow Lipinski rules and have a good partition coefficient. SwissADME predictions suggested that symmetrical phenylenediamine Schiff’s base derivatives have optimum parameters for anticancer activity and can be considered as lead molecules for further modifications
Molecular docking analysis
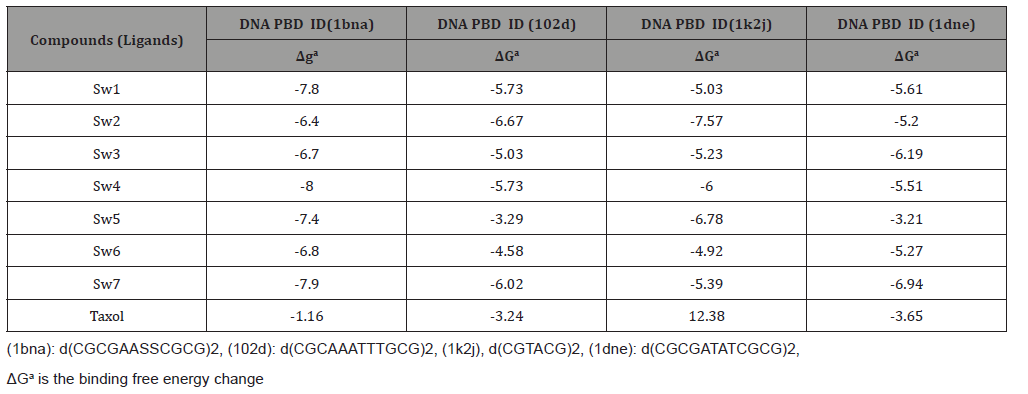
Table 1 shows the binding energies of the SW compounds and the DNAs fragments obtained by docking the investigated compounds within the DNA molecule. In this part of the current study, the molecular dockings of SW compounds with four B-DNA fragments were performed using Auto Dock 4.2 to understand the possible binding mode of SW compounds with B-DNA and to obtain information about interaction forces between ethidium bromide and DNA. Ethidium bromide and DNA were kept as flexible molecules and were docked into four forms of rigid B-DNA fragments to obtain the preferential binding site to SW compounds on B-DNAs. The molecular docking results are shown in (Table 1).
The molecular modeling results showed that there are van der Waals, hydrogen bonds and electrostatic interactions between ethidium bromide, SW compounds and DNA. The contribution of van der Waals and hydrogen bond interaction is much greater than that of the electrostatic interaction because the sum of van der Waals energy, hydrogen bonding energy and desolvation free energy is larger than the electrostatic energy, which is consistent with the experimental results shown in Table 2. As indicated in the literature, the binding mode of ethidium bromide on DNA is intercalating binding [28]. The fluorescence emission intensity of ethidium bromide decreased on addition of SW compounds suggesting that there is competitive binding between ethidium bromide and SW compounds on G-DNA. Therefore, it can be concluded that the binding mode of ethidium bromide on G-DNA is intercalating binding which is consistent with our modeling and fluorescence results (Table 1).
Table 1:Various energies in the binding process of the SW compounds with DNAs obtained from molecular docking. The unit of all energies was kcal/mol.
DNA binding properties
To study how proficiently the synthesized compounds, interact with G-DNA, the DNA binding primary properties were explored by using fluorescence emission spectroscopy. Fluorescence emission experiments have been extensively used to study the synthesized compound-DNA interactions, in which the fluorescence emissions of compounds (SW1-SW7) were increased significantly in the presence of the G-DNA. This is in good agreement with the fluorescence behavior of other intercalators reported in the literature [29]. The results indicated that all the compounds can interact with G-DNA.
Ethidium bromide (EB) competition assay
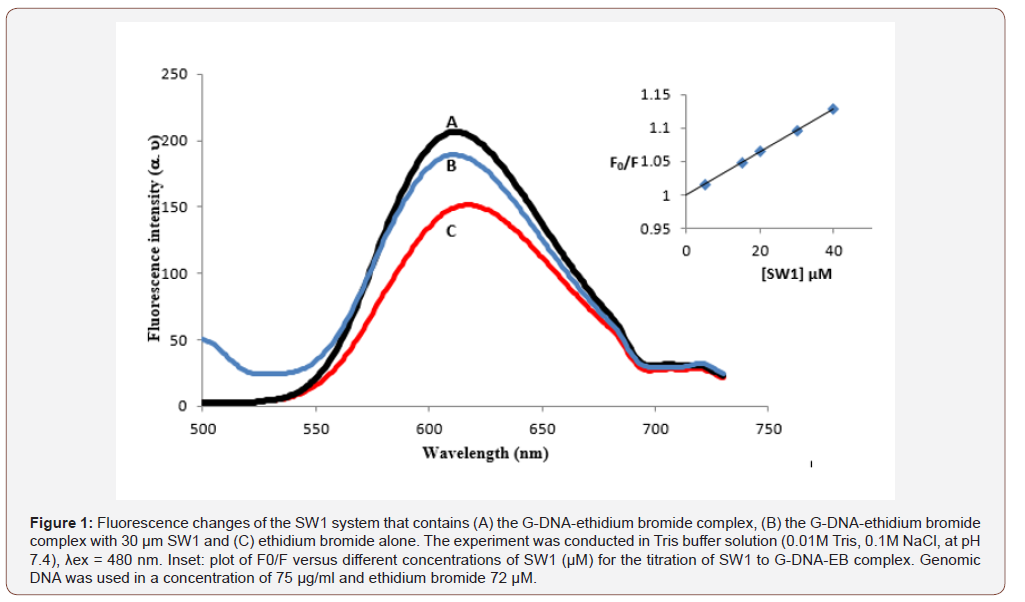
The structural properties of the complex formed between genomic DNA was studied using Raman microscopy equipped with near-infrared laser excitation [30]. The study showed that the phenanthridinium plane is tilted by 35 ± 5° from the plane perpendicular to the fiber (DNA helix) axis. The study also indicated that the coplanarity of the phenanthridinium ring and its immediate base neighbors at the intercalation site have a tilt angle closer A-DNA (20°) than to B-DNA (6°) and the average base tilt in stretches of DNA between intercalation sites remains to that of B-DNA [30]. The entrenched quenching assay based on the displacement of the intercalating dye, ethidium bromide, from G-DNA was utilized to explore the interaction mode between the complexes and G-DNA. EB is an extremely helpful DNA structural probe, which shows an important increase in fluorescence intensity when intercalating into the base pair of DNA. However, the increased fluorescence can be quenched obviously when there is a second complex that can replace the bound EB or break the secondary structure of DNA [31,32]. It has been shown that groove DNA binders can cause a decrease in EB emission intensity [33,34]. The entire compounds do not fluoresce with an excitation at 612 nm. The EB competition assay results are shown in Figure 1. The fluorescence intensity of DNA-bound EB at 612 nm decreased outstandingly with an increase of the compound (SW1–SW7) concentration. This decrease in fluorescence intensity may be due to the quenching of some EB molecules that were released from DNA into the solution after being substituted by the SW1–SW7 compounds. The phenomenon that the fluorescence of DNA-bound EB was quenched as a result of DNA and compound interactions is a typical sign of intercalation [33,34].
According to the literature the fluorescence quenching of DNA-bound EB can be well described by the linear Stern-Volmer equation [35-37] in which the synthesized (SW1–SW7) compounds were the quenchers: I0/I = 1 + KSV [Q]. I0 and I represent the fluorescence intensities in the absence and presence of quencher, respectively; KSV is a linear Stern-Volmer quenching constant; Q is the concentration of quencher. The KSV values were given by the ratio of the slope to intercept.
The KSV values for the (SW1–SW7) compounds are summarized in Table 2. As shown is Table 2, compound SW7 had the highest KSV value, which suggests that compound SW7 binds most strongly to G-DNA-even more than taxol (Figure 1) (Table 2).
Table 2:Some chemical, physical properties and the key selection vector (KSV) values of compounds (SW1–SW7).
Conclusion
We have synthesized seven symmetrical 1,2-phenylenediamine compounds and characterized using various spectroscopic techniques. DNA binding with SW compounds was investigated by fluorescence spectral techniques and revealed an intercalative interaction between the synthesized compounds and G-DNA. Among the investigated SW compounds, SW7 showed the highest binding affinity.
Acknowledgement
The authors gratefully acknowledge the technical support and valuable suggestions obtained from MS Halima Abdul Gbaj.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest and it is compliance with ethical standards. Ethical approval: This article does not contain any studies with human participants performed by any of the authors.
References
- Hartley JA, Hochhauser D (2012) Small molecule drugs-optimizing DNA damaging agent-based therapeutics. Curr Opin Pharmacol 12(4): 398-402.
- Srinivasan A, Gold B (2012) Small-molecule inhibitors of DNA damage-repair pathways: an approach to overcome tumor resistance to alkylating anticancer drugs. Future Med Chem 4(9): 1093-1111.
- Hamilton PL, Arya DP (2012) Natural product DNA major groove binders. Nat Prod Rep 29(2): 134-143.
- Denny WA (2001) DNA minor groove alkylating agents. Curr Med Chem 8(5): 533-544.
- Lucas MF, Cabeza DVI, Takahashi R, Rubio-Martinez J, Guallar V (2014) Atomic level rendering of DNA-drug encounter. Biophys J 106(2): 421-429.
- Xi H, Davis E, Ranjan N, Xue L, Hyde-Volpe D, et al. (2011) Thermodynamics of nucleic acid "shape readout" by an aminosugar. Biochemistry 50(42): 9088-9113.
- Fairbanks SD, Robertson CC, Keene FR, Thomas JA, Williamson MP (2019) Structural Investigation into the Threading Intercalation of a Chiral Dinuclear Ruthenium (II) Polypyridyl Complex through a B-DNA Oligonucleotide. J Am Chem Soc 141(11): 4644-4652.
- Bera D, Verdonck L, Glassner M, Madder A, Hoogenboom R (2019) Thermoresponsive DNA by Intercalation of dsDNA with Oligoethylene-Glycol-Functionalized Small-Molecule Intercalators. Macromol Rapid Commun 40(10): e1800900.
- Zanoza SO, Klimenko KO, Maltzev GV, Bykova TI, Levandovskiy IA, et al. (2019) Aminoalkoxyfluorenones and aminoalkoxybiphenyls: DNA binding modes. Bioorg Chem 86: 52-60.
- Schonn I, Hennesen J, Dartsch DC (2010) Cellular responses to etoposide: cell death despite cell cycle arrest and repair of DNA damage. Apoptosis 15(2): 162-172.
- Ross WE, Bradley MO (1981) DNA double-stranded breaks in mammalian cells after exposure to intercalating agents. Biochim Biophys Acta 654(1): 129-134.
- Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF (1984) Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 226(4673): 466-468.
- Gao Y, Li J, Huang G, Yan L, Dong Z (2015) Spectroscopic studies on the interaction between anthragallol and DNA using of ethidium bromide as a fluorescence probe. Spectrochim Acta A Mol Biomol Spectrosc 141: 239-243.
- Zhang G, Hu X, Fu P (2012) Spectroscopic studies on the interaction between carbaryl and calf thymus DNA with the use of ethidium bromide as a fluorescence probe. J Photochem Photobiol B 108: 53-61.
- Thies H, Schonenberger H, Bauer Kh (1958) Reactions of Schiff's bases. V. Transformation of benzylidene-arylamines with mixtures of magnesium and magnesium iodide. Arch Pharm Ber Dtsch Pharm Ges 219/63(11-12): 620-627.
- Puchtler H, Meloan SN, Brewton BR (1975) On the history of basic fuchsin and aldehyde-Schiff reactions from 1862 to 1935. Histochemistry 41(3): 185-194.
- Piscopo E, Diurno MV, Cirino G, Antonucci M, Cataldi MT et al. (1984) Biological activity of 4-hydroxy-5-formylbenzoic acid derivatives. II. Esters and Schiff bases with antimicrobial activity. Boll Soc Ital Biol Sper 60(3): 501-507.
- Lubec G, Leban J, Peyroux J, Sternberg M, Pollak A, et al. (1982) Reduced collagenolytic activity of rat kidneys with steptozotocin diabetes. Nephron 30(4): 357-360.
- Rahman AH, Ismail EM (1976) Synthesis of Schiff bases of benzofuran with potential biological activity. Arzneimittelforschung 26(5): 756-759.
- Lamie PF, Ali WAM, Bazgier V, Rarova L (2016) Novel N-substituted indole Schiff bases as dual inhibitors of cyclooxygenase-2 and 5-lipoxygenase enzymes: Synthesis, biological activities in vitro and docking study. Eur J Med Chem 123: 803-813.
- Meng XY, Liu JQ, Zhang XP, Chen XH, Yu AZ, et al. (1994) Synthesis and biological activities of 2,4-diamino-5-fluoro-6-substituted benzylamino quinazolines. Yao Xue Xue Bao 29(4): 261-267.
- Sofian S Mohamed, Inass A Al sadawi, Mohamed A Gbaj, Sami G Alsabri, Nahd M Elmaki, et al. (2018) Microwave Assisted Synthesis and Antimicrobial Evaluation of Symmetrical 1,2-Phenylenediamine Schiff's Base derivatives. Pharmacy & Pharmacology International Journal 6(5): 344-348.
- Johns MB, Paulus-Thomas JE (1989) Purification of human genomic DNA from whole blood using sodium perchlorate in place of phenol. Anal Biochem 180(2): 276-278.
- Daina A, Michielin O, Zoete V (2017) Swiss ADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7: 42717.
- Guan L, Yang H, Cai Y, Sun L, Di P et al. (2019) ADMET-score - a comprehensive scoring function for evaluation of chemical drug-likeness. Medchemcomm 10(1): 148-157.
- Yang H, Lou C, Sun L, Li J, Cai Y, et al. admet SAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019; 35(6):1067-1069.
- Tiwari R, Mahasenan K, Pavlovicz R, Li C, Tjarks W (2009) Carborane clusters in computational drug design: a comparative docking evaluation using AutoDock, FlexX, Glide, and Surflex. J Chem Inf Model 49(6): 1581-1589.
- Krishna AG, Kumar DV, Khan BM, Rawal SK, Ganesh KN (1998) Taxol-DNA interactions: fluorescence and CD studies of DNA groove binding properties of taxol. Biochim Biophys Acta 1381(1): 104-112.
- Kuruvilla E, Joseph J, Ramaiah D (2005) Novel bifunctional acridine-acridinium conjugates: synthesis and study of their chromophore-selective electron-transfer and DNA-binding properties. J Phys Chem B 109(46): 21997-22002.
- Tsuboi M, Benevides JM, Thomas GJ (2007) The Complex of Ethidium Bromide with Genomic DNA: Structure Analysis by Polarized Raman Spectroscopy. Biophys J 92(3): 928-934.
- Zhang QQ, Zhang F, Wang WG, Wang XL (2006) Synthesis, crystal structure and DNA binding studies of a binuclear copper (II) complex with phenanthroline. J Inorg Biochem 100(8): 1344-1352.
- Jiang M, Li YT, Wu ZY, Liu ZQ, Yan CW (2009) Synthesis, crystal structure, cytotoxic activities and DNA-binding properties of new binuclear copper(II) complexes bridged by N,N'-bis(N-hydroxyethylaminoethyl)oxamide. J Inorg Biochem 103(5): 833-844.
- Prunkl C, Pichlmaier M, Winter R, Kharlanov V, Rettig W, et al. (2010) Optical, redox, and DNA-binding properties of phenanthridinium chromophores: elucidating the role of the phenyl substituent for fluorescence enhancement of ethidium in the presence of DNA. Chemistry 16(11): 3392-3402.
- Garbett NC, Hammond NB, Graves DE (2004) Influence of the amino substituents in the interaction of ethidium bromide with DNA. Biophys J 87(6): 3974-3981.
- Li H, Yu YY, Hu X, Cao SW (2008) Research on the interactions between genistein and its glucosides with DNA. Guang Pu Xue Yu Guang Pu Fen Xi 28(8): 1905-1909.
- Yu YY, Li H, Hu X, Cao SW (2008) Research on the interaction of Cr (III) complex of genistein with DNA. Guang Pu Xue Yu Guang Pu Fen Xi 28(7): 1587-1591.
- Dezhampanah H, Fyzolahjani S (2013) Study on interaction of cationic porphyrazine with synthetic polynucleotides. Anal Cell Pathol (Amst) 36(5-6): 125-132.
-
Abdul M Gbaj, JSofian S Mohamed, Inass A Sadawe, Nisreen H Meiqal, Abdulathim A Alshoushan, Aisha S Aboud et al. DNA Interaction Study of Some Symmetrical 1,2-Phenylenediamine Schiff’s Base Derivatives as New Potential DNA Intercalators Using Ethidium Bromide Competition Fluorescent Assay. Arch Biomed Eng & Biotechnol. 2(2): 2019. ABEB.MS.ID.000534.
-
Fluorescence, DNA intercalation, Key selection vector, Double helix, Electronic interactions, Antiviral, Antibacterial, Anti-inflammatory, Antiproliferative, Antifungal, Antimalarial, Buffer solutions, Hybridization
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






