 Research Article
Research Article
Deep-Learning-Based Automated Quantification of Ventilation Defects on 3D Isotropic Hyperpolarized 129Xe Lung MRI
Tuneesh K Ranota1, Fumin Guo2, Ramtin Babaeipour1, Tingting Wu3, Matthew S Fox3,4 and Alexei Ouriadov1,3,4*
1School of Biomedical Engineering, Faculty of Engineering, The University of Western Ontario, London, Canada
2Sunnybrook Research Institute, University of Toronto, Toronto, Canada
3Department of Physics and Astronomy, The University of Western Ontario, London, Canada
4Lawson Health Research Institute, London, Canada
5Robarts Research Institute, London, ON, Canada
6Department of Medical Biophysics, The University of Western Ontario, London, ON, Canada
Alexei V Ouriadov, The University of Western Ontario, Canada.
Received Date: November 30, 2023; Published Date: January 02, 2024
Abstract
Semi-automated-segmentation quantifies non-isotropic Xenon-129 lung images in order to provide the Ventilation-Defect-Percent (VDP), but this method is not ideally suited for 3D-isotropic-voxel analysis. 3D-isotropic-voxel 129Xe images can be used to calculate semi-automated and deep learning-based automated VDP values for lung disease assessment. We aimed to develop a fully-automated Deep Learning-based (DL) segmentation algorithm for 3D 129Xe MRI analysis which generates VDP. This is a prospective study which included 10 participants (COVID-19 Survivors) with ventilation heterogeneity. A DL segmentation method was used to compute VDP and was compared to the semi-automated method serving as the benchmark reference. Imaging was acquired from Fast-Gradient-Recalled-Echo in 16sec breath-hold. Isotropic imaging was generated using a zerofilling approach. A non-significant mean difference for semi-automated-segmentation and DL-based VDP values was observed. SNR values were above 5 (Rose criteria).
The two VDP estimates had an intercept of -0.03, a slope of 1.1 and r=0.89. Bland Altman analysis indicated negligible bias and Sørensen- Dice (similarity) coefficients suggested a good match between the ground truth (semi-automated-segmentation) and DL segmentations. This study establishes the use of a deep-learning-based algorithm for segmentation and suggests the proposed method can be an alternative for time-consuming and higher variability segmentation methods.
Keywords:Lung; Deep Learning; Hyperpolarized Xenon-129 MRI; SARS-CoV-2 Infection; Ventilation Defects
Introduction
Hyperpolarized Xenon-129 magnetic resonance imaging (MRI) is an established research tool which has recently (December 2022) received the Food and Drug Administration (FDA) approval. The method provides sensitive and unique functional information in the lungs for observation and therapy guidance/assessment for individuals with respiratory conditions such as chronic obstructive pulmonary disease (COPD) [1,2], asthma [3], and Cystic Fibrosis (CF) [4,5]. Several recent studies have indicated that improved 129Xe polarization techniques have allowed for high spatial and temporal resolution pulmonary images, [2,6] that quantify ventilation heterogeneity and can provide calculation of ventilation defect percent (VDP) [7] for individuals diagnosed with COPD and asthma.
Recently, a new emerging respiratory disease caused by SARSCoV- 2, Coronavirus Disease 19 (COVID-19) [8,9], has influenced a pandemic of respiratory illness around the world with particular severe course in the elderly [10]. Symptoms of COVID-19 infection involve the upper respiratory tract and can vary from mild, such as the common cold, to severe, including pneumonia [11]. A recent study highlighted the importance of using 129Xe MRI to evaluate pulmonary function damage and microstructural parameters in COVID-19 participants, by finding significant differences in VDP [12]. In addition, studies have used hyperpolarized xenon-129 to identify long-term symptoms following COVID-19 infection, which Computed Tomography (CT) scans were unable to detect [13]. One study used 129Xe MRI to detect long-term pulmonary injury due to COVID-19 and found results which indicate compromised gas exchange in the lungs, which provide reasoning for symptoms that could not be understood through other imaging approaches [14].
Another study found objective damage in gas transfer in the lungs of COVID-19 participants with normal CT scans using 129Xe MRI [15]. These studies conclude that the use of 129Xe MRI in COVID studies will allow for an increased understanding of the symptoms after COVID-19, as it provides evidence of lung abnormalities that are not detected with conventional imaging [14,15]. Furthermore, studies are continuing to investigate the extent and repercussions of prolonged or extended symptoms of COVID-19 through the use of 129Xe MRI to improve lung disease progression [16].
Many studies quantifying ventilation heterogeneity in participants with pulmonary disease by calculating VDP have predominately used non-isotropic voxel scans [voxel size = 5x5mm2) 129Xe MRI datasets [2,17,18]. Newer non-Cartesian methods such as FLORET and 3D radial have also been used with non-isotropic voxels and are being investigated to improve 129Xe ventilation imaging and acquisition of isotropic resolution ventilation images have been demonstrated [19,20]. The development of high-resolution isotropic voxel in 129Xe imaging faces a number of obstacles such as insufficient, <5, [21] Signal-to Noise Ratio (SNR), [22,23] which does not allow for an accurate generation of the VDP estimates [24]. Another limitation is MRI scan time due to the relatively short breath-hold durations required to obtain the 3D isotropic-voxel 129Xe MRI static ventilation images. The 129Xe MRI modality has been found to be well tolerated by participants with lung diseases in general, [18,25- 27] but the breath-hold should be limited to approximately 16 seconds.
This allows for the acquisition of sixteen 15mm slices and collection of the non-isotropic voxel static ventilation images. High-resolution datasets include approximately 80 slices, thus requiring 80-seconds of breath-hold which is not physically possible from participants. The compressed sensing technique can help to minimize the scan time, however, the highest acceleration factor used for the 129Xe static ventilation imaging was three [28], while one needs 5 to finish the acquisition in 16 seconds.
Efforts have been undertaken to address the challenge of non-isotropic voxel limitations. In this context, isotropic-voxel high-resolution 3D 129Xe static-ventilation images were acquired through a single 16sec breath-hold using Fast Gradient Recalled Echo (FGRE), focusing on asthma subjects [22,24,29,30]. 129Xe high-resolution imaging is important as it should permit more accurate assessment of disease-progression, inform treatment options, and improve our understanding of ventilation heterogeneity. In our work, we acquired the non-isotropic 16-slice dataset using 129Xe FGRE MR imaging and generated the pseudo isotropic xenon 80-slice dataset using a zero-filling method.
The quantification of the isotropic-voxel high-resolution datasets can be challenging for several reasons. Presently used semi-automated segmentation [31] is able to provide ventilation-defect- percent estimates from 3D non-isotropic 129Xe lung images. Previous methods to quantify ventilation abnormalities include manual and semi-automated segmentation [31], the latter being the current gold standard. Algorithms to segment thoracic cavity images include seeded region-growing [31], clustering [32], and model-based techniques [33]. However, these methods are not optimal for isotropic-voxel high-resolution 3D 129Xe MRI analysis, partly due to the large number of slices [~80), thus creating a very time-consuming task. Recently, deep learning methods have been proposed to automatically segment the thoracic cavity region and ventilated region for non-isotropic 129Xe MRI [34].
Deep Learning (DL) is a type of machine learning that simulates human brain functions using multi-layered artificial neural networks. Its effectiveness hinges on two main factors: data availability and computational power. Recent advancements in data, computer processing, and graphics processors have significantly enhanced DL, leading to deeper and more successful neural networks. Convolutional neural networks (CNNs) leveraged by DL-based techniques have gained prominence in various medical imaging applications, particularly in image segmentation [35]. The primary objective of CNNs is to learn the feature maps of an image for the purpose of image classification. However, in the context of image segmentation, the aim shifts towards reconstructing an image from the feature map and assigning a classification to each individual pixel. Presently, DL approaches have shown a multitude of achievements in the field of medical image analysis [36] due to their efficiency and accuracy, such as brain tumor segmentation [37], lung segmentation in CT images [38], breast cancer radiotherapy [39] and for segmenting the lung to monitor potential biomarkers of pulmonary perfusion in COPD [40].
Segmentation models based on convolutional neural networks [41], such as U-Net [42] and U-Net++ [43], show potential to be used for VDP calculation. A study has demonstrated the use of a Pix2Pix generative adversarial network to generate synthetic 129Xe ventilation images that contain arbitrary ventilation defects and corresponding segmentation masks of the thoracic cavity [44]. Employing deep learning techniques such as CNN, has significantly enhanced pulmonary functional imaging by augmenting the quality of functional images, decreasing acquisition time and improve image segmentation and reconstruction [45-47]. Studies have used machine-learning algorithms in hyperpolarized gas MRI to predict lung ventilation heterogeneity in COPD participants [48] and to reconstruct human lung gas MRI from under-sampled k-space data [47]. We hypothesize that DL-based algorithms can be used for accurate generation of the VDP estimates from zero-filled generated isotropic-voxel images [80 slices] and can provide accurate assessment of lung structure and function.
In this study, 3D 129Xe static-ventilation data was acquired from ten COVID-19 survivors assuming that this lung disease causes ventilation defects [49]. The fully automated deep learning-based algorithm was used to generate VDP estimates from the isotropic voxel data, contrasting with a semi-automated method [31]. as the reference gold standard and/or ground truth. Our work on DL-based segmentation of hyperpolarized gas MRI distinguishes itself from prior studie by Tustison, et al. [50] and Astley, et al. [51] through the implementation of two distinct methodologies. Primarily, the U-Net++ architecture for segmentation was used, a deviation from the U-Net and nn_U-Net leveraged in earlier studies, respectively. As an advanced iteration of the U-Net architecture, U-Net++ incorporates supplementary skip connections and dense convolutional blocks, thereby boosting the precision of segmentation. Secondly, a transfer learning strategy was adopted from the ImageNet dataset, enabling us to fine-tune a pre-existing model to cater to our specific task with limited dataset.
This approach has been empirically demonstrated to bolster the performance of deep learning models across a variety of medical imaging tasks, including segmentation. The combination of these two techniques, namely the use of the advanced U-Net++ architecture and the application of transfer learning, not only differentiates our work from previous studies but also contributes a unique and valuable perspective to the field of hyperpolarized gas MRI segmentation. We believe that our methodology, which effectively balances computational efficiency with segmentation precision, offers a promising avenue for future research in this domain.
Materials and Methods
Study Participants
Ten COVID-19 Survivors were enrolled and provided written inform consent following the study protocol approved by a local ethics board.
Pulmonary Function Tests
Spirometry, plethsmography and the diffusing-capacity- of-the-lung-for-carbon-monoxide (DLco) were performed according to American Thoracic Society (ATS) guidelines [52] using a plethysmograph with an attached gas analyzer [MedGraphics Corporation. St. Paul, MN USA) to obtain the FEV1, forced vital capacity, residual volume and total lung capacity.
129Xe and 1H MRI Acquisition
129Xe MR imaging was performed at 3.0T (MR750, GEHC, WI) employing whole-body-gradients (Gmax=5 G/cm, slew rate=200 mTm- 1s-1), in accordance with previously described methods [53], and a commercial coil, 129Xe quadrature-flex RF[MR Solutions, USA). The 129Xe gas was polarized to 35% [initially 86% enriched) and was obtained from a turn-key, spin-exchange polarizer system (Polarean- 9820 129Xe-polarizer) [54]. A 30/70 by volume 129Xe/4He mixture of 1L was inhaled by all participants from functional residual capacity (FRC) during a 16 second breath-hold. Non-isotropic voxel xenon-static-ventilation images were obtained using a coronal- plane 3D FGRE sequence, TE/TR=1.5ms/5.1ms, reconstructed matrix size=128x128x16, initial flip angle=1.3o, FOV=40x40x- 24cm3, and voxel-size=3x3x15mm3, as previously described [7]. 1H MRI was performed as previously described [55]. The 128x128x80 slice datasets were generated using the 128x128x16 matrix size and then zero-filled 5-fold in the slice direction of the original 3D k-spaces.
Image Analysis
SNR Calculations
The SNR of 129Xe images was computed within three central coronal- view slices. This was achieved byusing a 15x15 voxel square region of interest situated within a lung area of homogeneous signal, along with the same 15x15 voxel square region of interest placed outside the lung in an area of devoid of lung signal [56].
VDP Calculation using a Semi-Automated method
Hyperpolarized gas MRI enables the observation of lung functionality by identifying unventilated areas within the lung, referred to as ventilation defects. The measurement of ventilation defects is performed using the Ventilation Defect Percent [VDP), which involves determining the ratio between the overall volume of ventilation defects [VDV) and the total volume of the thoracic cavity [TCV). This calculation applies to both semi-automated and fully automated deep learning techniques. The identification of ventilation defects was accomplished through the application of a k-means clustering methodology. For semi-automated segmentation, a k-means clustering algorithm previously described was used for semi-automated VDP calculations [30,31]. Two trained observers who completed 1-week full-time training with certification at Western University calculated the semi-automated-based VDP calculations, taking approximately 45-mins to 1-hour for each participant [80 slices each). Thus, taking approximately 10 hours per observer to obtain semi-automated VDP values.
VDP Calculation using a Fully-Automated approach
U-Net is a commonly used DL segmentation network which allows for fast and precise segmentation of images and has outperformed previous convolutional networks [41]. U-Net++ was introduced in the field to overcome problems of U-Net, such as the unknown optimal depth of encoder-decoder in each specific task and the restrictive design of skip connections. Our study used a semantic segmentation task [57]. Semantic segmentation involves assigning a class label to every pixel within an image. In our research we had two classes: 1) pixels belonging to the lung 2) pixels that do not belong to the lung area. In this work, we used U-Net++ composed of two pathways [43]. First, an encoder down-sampled the image while extracting the features using convolution and pooling layers. The objective in this instance was to encapsulate the context of the input image. Secondly, a decoder up-sampled the information from the encoder which resulted in an accurate localization.
At the same time, the encoder’s high-resolution contextual information was relayed to the decoder via skip connection to assist with localization. Finally, the contextual information from the encoder path was combined with the localization in the decoder to restore the size of the image and produce the predicted segmentations. In U-Net++, The redesigned skip connections have been introduced to decrease the semantic disparity between the contracting and expanding pathways, a significant improvement over the original U-Net architecture.
Since training a convolutional neural network with randomly initialized weights demands a substantial volume of data, transfer learning was used [58] in our architecture. ResNet 152 was utilized, which was trained on the ImageNet [59] dataset. ImageNet, encompassing over 14 million images, is frequently employed to train models for computer vision. This includes object categories such as animals, vehicles, and household objects. Training our model on such a big dataset gave us the advantage of introducing our network to basic and common concepts of images like edges and light intensity transitions. As a result, our network was pretrained on a bigger dataset to make it ready for our own MRI data. As the backpropagation take place to update the Deep Neural Network, the partial derivative will get either very small or very large, resulting in the vanishing/exploding gradient problem [60,61]. To overcome this challenge, ResNet [62] was proposed in 2015, introducing the concept of Residual Blocks combined with skip connection. Considering the advantages of this network, configuration of ResNet with 152 layers was used as the backbone of implementing the U-Net++ architecture.
The use of a pre-trained version of ResNet 152 speeded up the training process and results in faster convergence. To improve the model’s robustness and preprocess the dataset, various data augmentation [63] techniques were employed. This included pixel translation within a range of [-20, 20] in both X and Y axes to make the model translation invariant. Additionally, rotations between [-30°,30°] were applied to the input images to enhance robustness against rotational variances. Furthermore, size and intensity scaling between 0.8 and 1.2 were performed to achieve size invariance in the model. Finally, elastic deformation which has proved very useful in image segmentation tasks [42] was applied.
The training dataset consisted of 800 129Xe image sets. Moreover, the entire partially annotated training dataset was also used as the validation dataset during training. The test dataset consisted of 10 image sets from 10 subjects. To train the model, the Adam [64] optimizer was selected as the optimization algorithm. Finally, we optimized the network parameters over 200 epochs, with 100 updates each. For every update in each epoch, only 20 2D slices were utilized from the complete training dataset to adjust the network weights. A learning rate of 0.0001 was used as the step size and set the batch size to 20 to expedite the learning process. The network was implemented on the Keras 2.2.4 and Python 2.7 platforms, harnessing the computational power of an NVIDIA Tesla P100 Graphics Processing Unit [GPU) by NVIDIA Corp., based in Santa Clara, CA, USA. The deep learning-based algorithm used a U-Net++ network [43]. This trained network was used to segment the lung in the 80 slices 1H images for each participant. An affine and deformable registration from the NiftyReg package was employed. [http://cmictig. cs.ucl.ac.uk/wiki/index.php/NiftyReg) to align the 1H MR images with the 129Xe volumes.
The lung-segmentation was warped and the 129Xe image signal within these adjusted lung masks was automatically divided into 5 clusters, employing a 3D k-means clustering approach [31]. VDP was calculated by normalizing 129Xe ventilation-defects represented by the 1st cluster to the warped lung-masks [65]. Establishing the ground truth for texture analysis necessitates the process of registration. Previous methods include 3He MRI co-registered with Computed Tomography (CT) using rigid [66], affine [67] and deformable [68] techniques. More recently, an automated approach has been developed by registering both CT and 3He MRI to 1H MRI using NiftyReg affine and deformable registration tools [65]. The overall workflow of the deep learning-based segmentation begins with input images which are pre-processed in order to make the inputs ready for the network. Then, the model starts training. After each epoch, the model’s hyperparameter are updated to reduce the model loss. At the end the segmentation masks are provided as the output of the network.
Statistical Analysis
Pearson correlation coefficient was used to determine the relationship between semi-automated and DL-based VDP values. T-tests were performed using SPSS Statistics, V26.0 (SPSS Inc., Chicago, IL). Results were considered significant when the probability of two-tailed type I error (α) was less than 5% (p<.05). Bland-Altman (BA) was conducted for both semi-automated and DL-based VDP estimates. The Sørensen-Dice [similarity) coefficient (DSC) [69) was used for validation:
Here, ‘y’ represents the ground truth image while ‘y_pred’ refers to the prediction generated by our model.
DSC values can range from 0 to 1. A value of 0 implies no spatial intersection between the ground truth segmentation mask and the predicted mask, while a value of 1 signifies complete overlap. DSCs were computed for three distinct slices - anterior, central, and posterior.
Results
Table 1 provides an overview of the participant demographics and results from pulmonary function tests. Participant’s age ranged from 29 to 76, mean forced expiratory volume (FEV1) =79 and mean forced vital capacity (FVC) = 81. The diffusing capacity for carbon monoxide (DLco) ranged from 63% to 91% and the Residual Volume (RV) from 73% to 116% for five participants. DLco and RV for participants 1 to 5 had inaccurate data due to hardware issues and thus were not reported. Figure 1 shows coronal view for the traditional resolution voxel (3x3x15mm3) and anterior to posterior 129Xe MRI static-ventilation slices fora representative COVID-19 survivor. Images visualize ventilation defects and heterogeneity in the lungs. The calculated mean SNR values of the 3 central slices across all study participants ranged from 13 to 106.
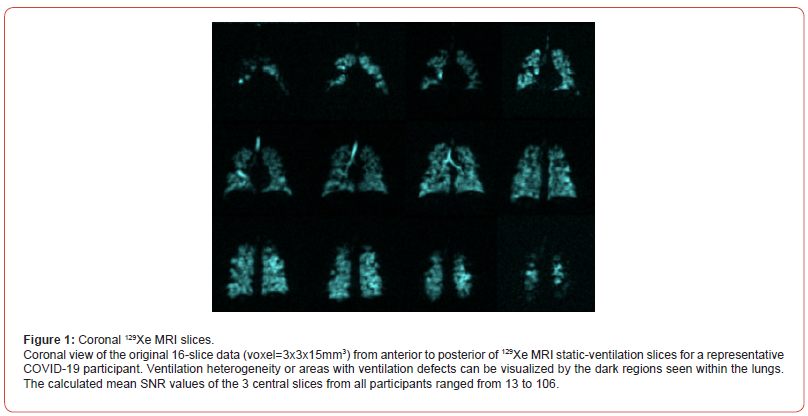
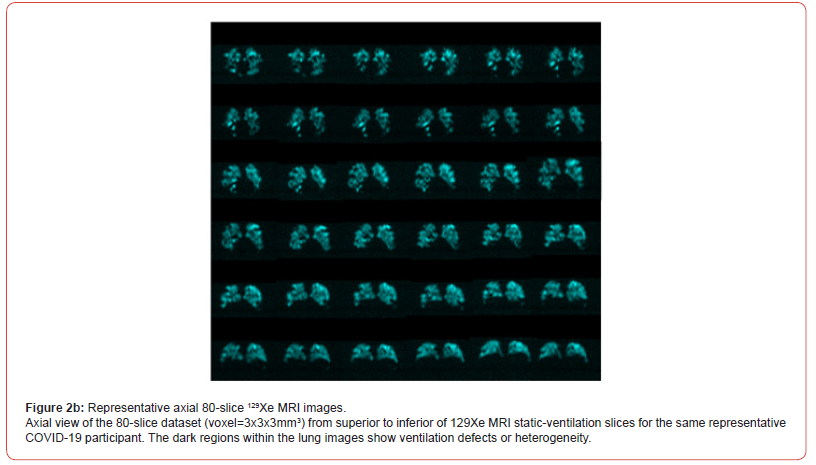
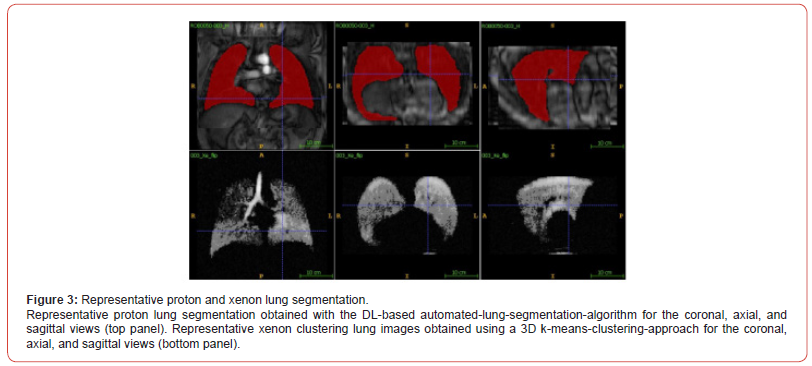
Figure 2a and 2b show coronal and axial view slices for the 80-slice dataset (voxel=3x3x3mm3) 129Xe MRI static-ventilation for same COVID-19 survivor, respectively. Images show ventilation defects and heterogeneity in the lungs. The calculated mean SNR values of the 3 central slices across all study participants ranged from 5.7 to 42.1 (Table 1). Figure 3 (top panel) shows proton lung segmentation obtained from the DL-based automated lung segmentation algorithm in coronal, axial and sagittal views. Figure 3 (bottom panel) displays the xenon lung segmentation obtained from the DLbased automated lung segmentation algorithm in coronal, axial and sagittal views after applying the k-means clustering approach in all three views.

The VDP values for the semi-automated segmentation method were 2.3%, 2.9%, 1.9%, 0.5%, 0.3%, 3.9%, 2.5%, 0.6%, 0.8%, 0.9% for participants 1 to 10, respectively (Table 1). The overall semi-automated VDP mean value was 1.7±0.72 (at 95% confidence interval). The VDP values for the deep learning-based segmentation method were 2.6%, 2.3%, 1.1%, 0.3%, 0.3%, 3.4%, 3.9%, 0.4%, 0.7%, 0.8% for participants 1 to 10, respectively (Table 1). The overall mean DL VDP value was 1.6±0.80 [at 95% confidence interval). Both semi-automated and deep learning-based VDP calculations were provided and the largest disagreement between two VDP estimates was found for COVID survivor-7, semi-automated VDP = 2.5% and DL VDP = 3.9%. This participant also showed the smallest SNR values.
Ventilation defect percentage for semi-automated and deep learning methods both showed a significant positive correlation with FEV1% [r = 0.72, P = 0.02 and r = 0.68, P = 0.3, respectively). Both semi-automated and DL-based VDP values did not correlate significantly with FVC%, RV% and DLco%. Figure 4 illustrates the correlation between VDP values obtained through semi-automated methods and those acquired via fully automated deep learning from all COVID-19 survivors: intercept=-0.03, slope=1.1, and r=0.89. Figure 5 shows the BA analysis for the semi-automated and deep learning-based VDP estimates and plot indicates the average of the two VDPs [solid line) and the 95% limits of agreement (dotted lines). Participants = 10, mean = -1.48±0.83, std. error mean = 0.26, r=0.51, R2=0.26, lower limit = -0.75 and upper limit = 0.45.
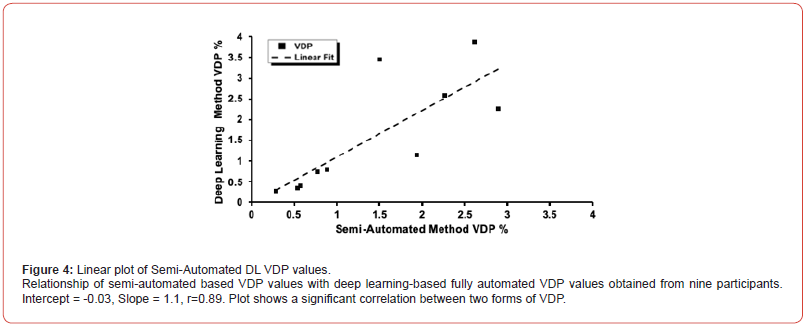
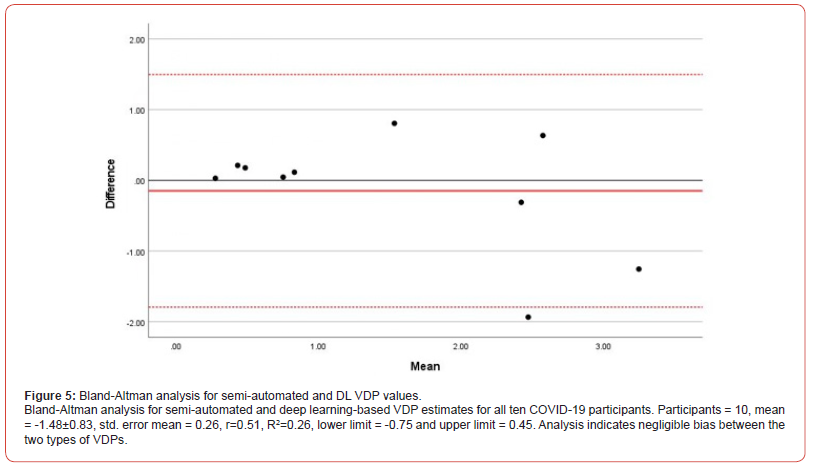
Table 1:Demographics and 129Xe MRI Results.

DSCs calculated for three central slices ranged from 91% to 96% (Table 1), the overall mean values were 94±1.0%, 93±1.0% and 94±1.2% for the three central slices, respectively. DSCs calculated for three anterior slices ranged from 54% to 93%, excluding 0 values, and overall mean values were 66.4±7.8%, 82.4±2.8% and 86.9±3.2%, for each slice. The DSCs calculated for three posterior slices ranged from 71% to 96%, excluding 0 values, and overall mean values were 91.3±1.2%, 68.9±11.1%, and 61.38±12.6%, for each slice.
Discussion
In this study, the potential for achieving complete automated segmentation of the 80-slice 129Xe lung images was demonstrated. The primary objective of this work was to apply deep learning- based algorithms to the 80-slice datasets and assess the ventilation heterogeneity within the lungs. The semi-automated lung segmentation method takes quite long [~45min to an hour per study subject) and should be replaced with more efficient approach to analyze the 80-slice datasets obtained from potentially larger participant studies. We believe that the quantitative analysis of the 129Xe lung images with better resolution may improve sensitivity to detect smaller ventilation deficits and therefore, more reliable for disease progression observation and therapy assessment. This aspect becomes significant in view of the FDA approval acquired for 129Xe MRI.
Ventilation defect percentage for semi-automated and deep learning methods showed a significant positive correlation with FEV1% [r = 0.7232, P = 0.01809 and r = 0.6821, P = 0.02978, respectively). Both semi-automated and DL-based VDP values did not correlate significantly with FVC%, RV% and DLco%. These results are expected, as previous studies, 129Xe ventilation MRI has found similar significant correlations between semi-automated VDP results and FEV1%, but no significant correlations with FVC and DLco for participants with asthma [22,70-72]. Another study found stronger correlations between FEV1% and 129Xe MRI-based VDP compared to phase resolved functional lung MRI in participants with CF [73]. Thus, it can be concluded that our correlation to FEV1% and VDP estimates are similar to previously reported literature.
SNR was calculated for both 16-slice datasets [original data] and 80-slice datasets [generated data] 129Xe MRI static-ventilation slices and were all above the Rose criteria of SNR=5 [21]. The SNR estimates for the 80-slice images were approximately half of the 16-slice images, but with voxel-size five times smaller [Table 1).
The smallest SNR value for both 16-slice and 80-slice images was found for COVID survivor 7, with SNR between 11 to 13 and 5.7 to 6.8, respectively. Previous studies have assessed the impact of SNR on DL-generated VDPs and that it is invariant to SNR with no significant impact on VDP accuracy [34,49]. This result demonstrates that the used zero-filling method generated sufficient quality 80-slice lung images. The sufficient SNR values of the 80-slice images permitted to conduct quantitative analysis and specifically, the semi-automated [31] and deep learning-based VDP calculations for each study participant. We used the current reference standard [semi-automated segmentation] to validate the deep learning- based approach conducting the lung segmentation and then the VDP estimate calculation utilizing the k-mean clustering method. To our knowledge this is the first report of the VDP estimates obtained from 80-slice COVID-19 survivors 129Xe lung images. The 80-slice 129Xe MRI-based VDP values have been previously measured in asthma participants using a high resolution data acquisition method and our mean VDP values reported for both semi-automated and deep learning methods were consistent with these results [22].
This further supports our hypothesis that DL-based algorithms can provide accurate VDP estimates and therefore will be able to provide accurate assessment of lung structure and function. A strong linear correlation between semi-automated and DL-based VDP values was found with a Pearson correlation coefficient of r=0.89 [the intercept was close to zero and slope close to unity, Figure 4 suggesting the reasonable preciseness of the VDP estimates obtained with the DL-based approach. COVID survivor 7 demonstrated the largest disagreement between the two VDP estimates obtained with two different approaches. This discrepancy may have resulted from low SNR values, suggesting that SNR values well above the Rose criteria (SNR=5) [21] should be used for accurate VDP estimation.
Furthermore, Bland Altman analysis indicated a negligible bias between the two types of VDPs. The t-score test was not statistically significant, Sig = 0.369, suggesting that there is no proportional bias. Bland Altman analysis assists in evaluating the level of agreement between two measurement methods [74], thus it can be concluded from these results that semi-automated and DL-based VDP calculations are both reliable and accurate. The two outliers seen in Figures 4 and 5 may have resulted from the combination of lower SNR and small training dataset. A smaller training dataset could potentially result in outliers due to overfitting, a scenario where the model excessively learns the training data, incorporating its noise or outliers. This overfitting can result in unexpected results when the model is applied to new, unseen data. Therefore, the outliers observed in Figures 4 and 5 may be a consequence of the combination of lower Signal-to-Noise Ratio (SNR) and the limited size of our training dataset.
The overall mean DSC values for slice 1 was 94 ± 1.0%, slice 2 was 93 ±1.0% and slice 3 was 94 ± 1.2% [at 95% confidence intervals). DSCs acquired from semi-automated and DL based VDP values are at a range of good reproducibility and indicate high spatial overlap between the two segmentation results [69]. Thus, suggesting a good match between the semi-automated and DL segmentations, further supporting the accuracy of DL-based VDP calculations.
This study used U-Net++ composed of two pathways [42]. In the U-Net++ architecture, the fundamental encoder-decoder structure is preserved, while enhancements are introduced through the incorporation of additional up-sampling and skip connections. These modifications facilitate the creation of U-Nets with varying depths. The central incentive for reconfiguring the skip connections was to lessen the semantic disparity between the contracting and expanding pathways., thereby simplifying the optimization process. Furthermore, U-Net++ introduces the concept of deep supervision. This innovative feature allows for the adjustment of model complexity, thereby striking a balance between computational speed and performance. The nested design enables the reuse of features from lower layers, effectively reducing the semantic disparity between feature maps at different scales. Simultaneously, the dense skip connections promote superior feature fusion and help alleviate the vanishing-gradient problem, a common issue in deep learning models.
Consequently, U-Net++ has showcased exceptional performance in a range of tasks, notably in medical image segmentation and object detection, often outperforming the original U-Net and other comparable architectures [75]. In this pilot study, several study limitations are acknowledged including low SNR-based errors, small number of study subjects, impossibility to acquire the 80 slice images, small dataset used for the network training, and using to different semi-automated segmentation methods for the data generation for the training network and ground truth. Additionally, the range of VDP values in the dataset are relatively small and thus may limit applicability of the proposed approach and DSC values on anterior and posterior slices tend to be much lower when compared to central slices.
SNR that is less than or around the Rose criteria is considered a limitation any quantitate analysis and therefore, for the VDP calculations. However, newer polarization methods with increased 129Xe polarization levels (400ml volume in 15-20 minutes providing ~50% polarization) can improve SNR and image quality. Enhanced progression of MRI hardware encompassing the integration of a rigid and more homogenous coil [53,76] along with a phased-receive- array [77] holds the potential to significantly enhance the 80-slice image quality and potentially decreasing dose of the isotopically- enriched 129Xe, and consequently, reducing the cost of 129Xe MRI for participants without compromising the image quality.
In addition, this study was limited by sample sizes of the participants, as only 10 participants with recent infection of COVID-19 were examined. However, the goals of this study were a calculation of the VDP estimates using two different methods (semi-automated and DL) using the generated 80-slice 129Xe lung images. Another limitation is a generation of 80-slice images using a zero-filling approach. The “gained” resolution that is obtained is an “apparent” resolution because it was acquired from a zero-filling interpolation routine. This is a limitation as it can provide implications for the detection of ventilation defects. Finally, it is worth mentioning that to create a more solid deep learning-based segmentation framework for the VDP calculation, it would be beneficial to test different segmentation architectures and compare them to select the best one that suits our data. To enhance our segmentation performance, we plan to experiment with various neural network architectures while also expanding the dataset size.
Additionally, in this research study manual segmentation dataset was used to train the neural network. However, since the semi-automated segmentation is the current golden standard and the final results are compared with that method, the semi-automated segmentation outputs will be used as the training data for our future works. Furthermore, we recognize that inter-observer variability between manual or semi-automated segmentations may be demonstrated and can be a limitation of this work.
Conclusion
The method of semi-automated-lung-segmentation is widely employed in hyperpolarized-gas lung imaging and for calculating VDP. However, the 80-slice dataset requires a significant observer time (~45min to an hour). By acquiring VDP estimates using deep learning-based algorithms, we increase time efficiency and reduce possibilities of human error. This study demonstrated that deep learning-based algorithms on 129Xe 80-slice static-ventilation imaging of COVID-19 survivors provides a way to calculate time-efficient VDP estimates, which allows for rapid evaluation of ventilation heterogeneity. This means that semi-automated segmentation methods can be potentially replaced by fully automated methods.
Funding Information
This research was funded by NSERC DG (R5942A04) and Western Research Catalyst Grant [R5942A03].
Author Contributions
Conceptualization, A.O.; Methodology, A.O., G.P., M.F., T.R., R.B., and T.W.; Software, F.G., T.R., R.B., and A.O.; Validation, A.O., T.R., and R.B.; Formal Analysis, A.O., T.R., and R.B.; Investigation, A.O., T.R., R.B., and T.W.; Resources, A.O., G.P., F.G., and M.F.; Data Curation, A.O., T.R. and R.B.; Writing – Original Draft Preparation, A.O., T.R., and R.B.; Writing – Review & Editing, A.O., T.R., and R.B.; Visualization, A.O., G.P., and T.R.; Supervision, A.O., Project Administration, A.O.; Funding Acquisition, A.O.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Review Board (Robarts Research Institute) [116775 November 03, 2020].
Consent Statements
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the participants to publish this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kirby M, Pike D, Sin DD, Coxson HO, McCormack DG, et al. (2015) COPD: Do Imaging Measurements of Emphysema and Airway Disease Explain Symptoms and Exercise Capacity? Radiology 277(3): 872-880.
- Kirby M, Svenningsen S, Owrangi A, Wheatley A, Farag A, et al. (2012) Hyperpolarized 3He and 129Xe MR imaging in healthy volunteers and patients with chronic obstructive pulmonary disease. Radiology 265(2): 600-610.
- Svenningsen S, Eddy RL, Lim HF, Cox PG, Nair P, et al. (2018) Sputum Eosinophilia and Magnetic Resonance Imaging Ventilation Heterogeneity in Severe Asthma. American journal of respiratory and critical care medicine 197(7): 876-884.
- Kołodziej M, de Veer MJ, Cholewa M, Egan GF, Thompson BR (2017) Lung function imaging methods in Cystic Fibrosis pulmonary disease. Respiratory research 18(1): 96-100.
- Walkup LL, Thomen RP, Akinyi TG, Watters E, Ruppert K, et al. (2016) Feasibility, tolerability and safety of pediatric hyperpolarized (129) Xe magnetic resonance imaging in healthy volunteers and children with cystic fibrosis. Pediatr Radiol 46(12): 1651-1662.
- Moller HE, Chen XJ, Saam B, Hagspiel KD, Johnson GA, et al. (2002) MRI of the lungs using hyperpolarized noble gases. Magn Reson Med 47(6): 1029-1051.
- Svenningsen S, Kirby M, Starr D, Leary D, Wheatley A, et al. (2013) Hyperpolarized (3) He and (129) Xe MRI: differences in asthma before bronchodilation. J Magn Reson Imaging 38(6): 1521-1530.
- Yuki K, Fujiogi M, Koutsogiannaki S (2020) COVID-19 pathophysiology: A review. Clinical immunology (Orlando, Fla) 215(1): 108427-108430.
- Velavan TP, Meyer CG (2020) The COVID-19 epidemic. Tropical medicine & international health: TM & IH 25(3): 278-280.
- Wu Z, McGoogan JM (2020) Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. Jama 323(13): 1239-1242.
- Su S, Wong G, Shi W, Liu J, Lai ACK, et al. (2016) Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends in microbiology 24(6): 490-502.
- Li H, Zhao X, Wang Y, Lou X, Chen S, et al. (2021) Damaged lung gas exchange function of discharged COVID-19 patients detected by hyperpolarized (129)Xe MRI. Science advances 7(1): eabc8180-eabc8185.
- Grist JT, Chen M, Collier GJ, Raman B, Abueid G, et al. (2021) Hyperpolarized (129)Xe MRI Abnormalities in Dyspneic Patients 3 Months after COVID-19 Pneumonia: Preliminary Results. Radiology 301(1): E353-E360.
- Dietrich O (2021) Detecting COVID-19-related Chronic Pulmonary Injury with (129)Xe MRI. Radiology 301(1): E373-e374.
- Grist JT, Collier GJ, Walters H, Chen M, Eid GA, et al. (2022) The Investigation of Pulmonary Abnormalities using Hyperpolarised Xenon Magnetic Resonance Imaging in Patients with Long-COVID. medRxiv: the preprint server for Health Sciences pp. 1-15.
- Wild JM, Porter JC, Molyneaux PL, George PM, Stewart I, et al. (2021) Understanding the burden of interstitial lung disease post-COVID-19: the UK Interstitial Lung Disease-Long COVID Study (UKILD-Long COVID). BMJ Open Respiratory Research 8(1): e001049-e001055.
- Westcott A, Guo F, Parraga G, Ouriadov A (2019) Rapid single-breath hyperpolarized noble gas MRI-based biomarkers of airspace enlargement. J Magn Reson Imaging 49(6): 1713-1722.
- Parraga G, Ouriadov A, Evans A, McKay S, Lam WW, et al. (2007) Hyperpolarized 3He ventilation defects and apparent diffusion coefficients in chronic obstructive pulmonary disease: preliminary results at 3.0 Tesla. Investigative radiology 42(6): 384-391.
- Willmering MM, Niedbalski PJ, Wang H, Walkup LL, Robison RK, et al. (2020) Improved pulmonary (129) Xe ventilation imaging via 3D-spiral UTE MRI. Magn Reson Med 84(1): 312-320.
- He M, Wang Z, Rankine L, Luo S, Nouls J, et al. (2020) Generalized Linear Binning to Compare Hyperpolarized (129)Xe Ventilation Maps Derived from 3D Radial Gas Exchange Versus Dedicated Multislice Gradient Echo MRI. Acad Radiol 27(8): e193-e203.
- Rose A (1948) The sensitivity performance of the human eye on an absolute scale. Journal of the Optical Society of America 38(2): 196-208.
- Barker AL, Westcott AR, Eddy RL, McCormack DG, Parraga G, et al. (2019) Feasibility of Single Breath-hold Isotropic Voxel 129Xe MRI in Patients ISMRM 19th Annual Meeting, Paris, France.
- Ranota TK, Serrai H, McCormack DG, Parraga G, Ouriadov A (2021) Feasbility of Single Breath-hold Isotropic Voxel 129Xe MRI in COVID-19 Survivors using a Key-Hole Method [abstract]. ISMRM 30th Annual Meeting, United States.
- Wolber J, Cherubini A, Dzik Jurasz AS, Leach MO, Bifone A (1999) Spin-lattice relaxation of laser-polarized xenon in human blood. Proc Natl Acad Sci USA 96(7): 3664-3669.
- Driehuys B, Martinez Jimenez S, Cleveland ZI, Metz GM, et al. (2012) Chronic obstructive pulmonary disease: safety and tolerability of hyperpolarized 129Xe MR imaging in healthy volunteers and patients. Radiology 262(1): 279-289.
- Shukla Y, Wheatley A, Kirby M, Svenningsen S, Farag A, et al. (2012) Hyperpolarized 129Xe magnetic resonance imaging: tolerability in healthy volunteers and subjects with pulmonary disease. Acad Radiol 19(8): 941-951.
- Lutey BA, Lefrak SS, Woods JC, Tanoli T, Quirk JD, et al. (2008) Hyperpolarized 3He MR imaging: physiologic monitoring observations and safety considerations in 100 consecutive subjects. Radiology 248(2): 655-661.
- Collier GJ, Hughes PJC, Horn FC, Chan HF, Tahir B, et al. (2019) Single breath-held acquisition of coregistered 3D (129) Xe lung ventilation and anatomical proton images of the human lung with compressed sensing. Magn Reson Med 82(1): 342-347.
- Norquay G, Collier GJ, Rao M, Maunder A, Rodgers OI, et al. (2017) Large-scale production of highly-polarized 129Xe. Proc 25th Annual Meeting ISMRM, Honolulu pp. 2140-2147.
- Ranota TK, Guo F, Wu T, Fox MS, Ouriadov A (2022) Automated Quantification of Ventilation Defects and Heterogeneity in 3D Isotropic 129Xe MRI. (Abstract) ISMRM 2022 Annual Meeting, London, England, UK.
- Kirby M, Heydarian M, Svenningsen S, Wheatley A, McCormack DG, et al. (2012) Hyperpolarized 3He magnetic resonance functional imaging semiautomated segmentation. Acad Radiol 19(2): 141-152.
- Coleman GB, Andrews HC (1979) Image segmentation by clustering. Proceedings of the IEEE 67(1): 773-785.
- McInerney T, Terzopoulos D (1996) Deformable models in medical image analysis: a survey. Med Image Anal 1(2): 91-108.
- Astley JR, Biancardi AM, Marshall H, Hughes PJC, Collier GJ, et al. (2023) A Dual-Channel Deep Learning Approach for Lung Cavity Estimation From Hyperpolarized Gas and Proton MRI. J Magn Reson Imaging 57(6): 1878-1890.
- Lei T, Wang R, Wan Y, Du X, Meng H, et al. (2020) Medical Image Segmentation Using Deep Learning: A Survey. arXiv pp. 1-23.
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, et al. (2017) A survey on deep learning in medical image analysis. Med Image Anal 42(5): 60-88.
- Ranjbarzadeh R, Bagherian Kasgari A, Jafarzadeh Ghoushchi S, Anari S, Naseri M, et al. (2021) Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images 11(1): 1-17.
- Zhao T, Gao D, Wang J, Yin Z (2018) Lung segmentation in CT images using a fully convolutional neural network with multi-instance and conditional adversary loss. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018): IEEE pp. 505-509.
- Men K, Zhang T, Chen X, Chen B, Tang Y, et al. (2018) Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med 50(1): 13-19.
- Winther HB, Gutberlet M, Hundt C, Kaireit TF, Alsady TM, et al. (2020) Deep semantic lung segmentation for tracking potential pulmonary perfusion biomarkers in chronic obstructive pulmonary disease (COPD): The multi‐ethnic study of atherosclerosis COPD study 51(2): 571-579.
- Shelhamer E, Long J, Darrell T (2017) Fully Convolutional Networks for Semantic Segmentation. IEEE transactions on pattern analysis and machine intelligence 39: 640-651.
- Ronneberger O, Fischer P, Brox T (2015) Convolutional Networks for Biomedical Image Segmentation.
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2020) UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans Med Imaging 39: 1856-1867.
- Leewiwatwong S, Lu J, Zhang J, Mummy D, Dummer I, et al. (2022) Ventilation Defect Synthesis in Hyperpolarized 129Xe Ventilation MRI to Accelerate Training of Segmentation Models. ISMRM.
- Ohno Y, Seo JB, Parraga G, Lee KS, Gefter WB, et al. (2021) Pulmonary Functional Imaging: Part 1-State-of-the-Art Technical and Physiologic Underpinnings. Radiology 299: 508-523.
- Park B, Park H, Lee SM, Seo JB, Kim N (2019) Lung Segmentation on HRCT and Volumetric CT for Diffuse Interstitial Lung Disease Using Deep Convolutional Neural Networks. Journal of digital imaging 32: 1019-1026.
- Duan C, Deng H, Xiao S, Xie J, Li H, et al. Fast and accurate reconstruction of human lung gas MRI with deep learning. Magn Reson Med 82(6): 2273-2285.
- Westcott A, Capaldi DPI, McCormack DG, Ward AD, Fenster A, et al. (2019) Chronic Obstructive Pulmonary Disease: Thoracic CT Texture Analysis and Machine Learning to Predict Pulmonary Ventilation. Radiology 293: 676-684.
- Matheson AM, McIntosh MJ, Kooner HK, Lee J, Desaigoudar V, et al. (2022) Never-hospitalized patients with post-acute COVID19 syndrome: 129Xe MRI gas-exchange abnormalities. British Medical Journal: Open Respiratory Research.
- Tustison NJ, Avants BB, Lin Z, Feng X, Cullen N, et al. (2019) Convolutional Neural Networks with Template-Based Data Augmentation for Functional Lung Image Quantification. Acad Radiol 26: 412-423.
- Astley JR, Biancardi AM, Hughes PJC, Marshall H, Smith LJ, et al. (2022) Large-scale investigation of deep learning approaches for ventilated lung segmentation using multi-nuclear hyperpolarized gas MRI. Sci Rep 12(1): 10566.
- Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, et al. (2005) Standardisation of spirometry. The European respiratory journal 26: 319-338.
- Ouriadov A, Farag A, Kirby M, McCormack DG, Parraga G, et al. (2013) Lung morphometry using hyperpolarized (129) Xe apparent diffusion coefficient anisotropy in chronic obstructive pulmonary disease. Magn Reson Med 70: 1699-1706.
- Kaushik SS, Cleveland ZI, Cofer GP, Metz G, Beaver D, et al. (2011) Diffusion-weighted hyperpolarized 129Xe MRI in healthy volunteers and subjects with chronic obstructive pulmonary disease. Magn Reson Med 65: 1154-1165.
- Kirby M, Pike D, Coxson HO, McCormack DG, Parraga G (2014) Hyperpolarized (3)He ventilation defects used to predict pulmonary exacerbations in mild to moderate chronic obstructive pulmonary disease. Radiology 273: 887-896.
- Thomen RP, Sheshadri A, Quirk JD, Kozlowski J, Ellison HD, et al. (2015) Regional ventilation changes in severe asthma after bronchial thermoplasty with (3)He MR imaging and CT. Radiology 274(1): 250-259.
- Brock KK (2010) Results of a multi-institution deformable registration accuracy study (MIDRAS). Int J Radiat Oncol Biol Phys 76(2): 583-596.
- Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, et al. (2004) Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Academic radiology 11(2): 178-189.
- Ebner L, Virgincar RS, He M, Choudhury KR, Robertson SH, et al. (2019) Multireader Determination of Clinically Significant Obstruction Using Hyperpolarized (129)Xe-Ventilation MRI. AJR Am J Roentgenol 212(4): 758-765.
- Roach D, Walkup L, Hall C, Thomen RP, McCormack FX, et al. (2018) Quantification of Cyst Ventilation Using Hyperpolarized <sup>129</sup>Xe MRI Correlates with Pulmonary Function Tests in Patients with Lymphangioleiomyomatosis. C97 DIFFUSE PARENCHYMAL LUNG DISEASES: EVALUATION, OUTCOMES, AND TRIALS: American Thoracic Society pp. A5924-A5924.
- Roach D, Walkup L, Hall C, Thomen R, McCormack F, et al. (2018) Quantification of cyst ventilation using hyperpolarized 129Xe MRI Correlates with pulmonary function tests in patients with lymphangioleiomyomatosis. C97 DIFFUSE PARENCHYMAL LUNG DISEASES: EVALUATION, OUTCOMES, AND TRIALS: American Thoracic Society pp. A5924-A5924.
- Couch MJ, Munidasa S, Rayment JH, Voskrebenzev A, Seethamraju RT, et al. (2021) Academic Radiology 28(8): e209-e218.
- Giavarina D (2015) Understanding bland altman analysis. Biochemia medica 25(2): 141-151.
- Walsh J, Othmani A, Jain M, Dev S (2022) Using U-Net network for efficient brain tumor segmentation in MRI images. Healthcare Analytics 2(1): 100098-100102.
- Farag A, Wang J, Ouriadov A, Parraga G GS (2012) Unshielded and asymmetric RF transmit coil for hyperpolarized 129Xe human lung imaging at 3.0 T. In Proceedings of the 20th Annual Meeting of ISMRM, Melbourne, Australia pp. 1233-1240.
- Chang YV, Quirk JD, Yablonskiy DA (2015) In vivo lung morphometry with accelerated hyperpolarized 3He diffusion MRI: A preliminary study. Magnetic Resonance in Medicine 73(4): 1609-1614.
- Thomen RP, Sheshadri A, Quirk JD, Kozlowski J, Ellison HD, et al. (2015) Regional ventilation changes in severe asthma after bronchial thermoplasty with (3)He MR imaging and CT. Radiology 274(1): 250-259.
- Brock KK (2010) Results of a multi-institution deformable registration accuracy study (MIDRAS). Int J Radiat Oncol Biol Phys 76(2): 583-596.
- Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, et al. (2004) Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Academic radiology 11(2): 178-189.
- Ebner L, Virgincar RS, He M, Choudhury KR, Robertson SH, et al. (2019) Multireader Determination of Clinically Significant Obstruction Using Hyperpolarized (129)Xe-Ventilation MRI. AJR Am J Roentgenol 212(4): 758-765.
- Roach D, Walkup L, Hall C, Thomen RP, McCormack FX, et al. (2018) Quantification of Cyst Ventilation Using Hyperpolarized <sup>129</sup>Xe MRI Correlates with Pulmonary Function Tests in Patients with Lymphangioleiomyomatosis. C97 DIFFUSE PARENCHYMAL LUNG DISEASES: EVALUATION, OUTCOMES, AND TRIALS: American Thoracic Society pp. A5924-A5924.
- Roach D, Walkup L, Hall C, Thomen R, McCormack F, et al. (2018) Quantification of cyst ventilation using hyperpolarized 129Xe MRI Correlates with pulmonary function tests in patients with lymphangioleiomyomatosis. C97 DIFFUSE PARENCHYMAL LUNG DISEASES: EVALUATION, OUTCOMES, AND TRIALS: American Thoracic Society pp. A5924-A5924.
- Couch MJ, Munidasa S, Rayment JH, Voskrebenzev A, Seethamraju RT, et al. (2021) Academic Radiology 28(8): e209-e218.
- Giavarina D (2015) Understanding bland altman analysis. Biochemia medica 25(2): 141-151.
- Walsh J, Othmani A, Jain M, Dev S (2022) Using U-Net network for efficient brain tumor segmentation in MRI images. Healthcare Analytics 2(1): 100098-100102.
- Farag A, Wang J, Ouriadov A, Parraga G GS (2012) Unshielded and asymmetric RF transmit coil for hyperpolarized 129Xe human lung imaging at 3.0 T. In Proceedings of the 20th Annual Meeting of ISMRM, Melbourne, Australia pp. 1233-1240.
- Chang YV, Quirk JD, Yablonskiy DA (2015) In vivo lung morphometry with accelerated hyperpolarized 3He diffusion MRI: A preliminary study. Magnetic Resonance in Medicine 73(4): 1609-1614.
-
Tuneesh K Ranota, Fumin Guo, Ramtin Babaeipour1, Tingting Wu, Matthew S Fox and Alexei Ouriadov*. Deep-Learning- Based Automated Quantification of Ventilation Defects on 3D Isotropic Hyperpolarized 129Xe Lung MRI. Arch Biomed Eng & Biotechnol. 7(4): 2024. ABEB.MS.ID.000670.
-
Lung; Deep Learning; Hyperpolarized Xenon-129 MRI; SARS-CoV-2 Infection; Ventilation Defects
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






