 Research Article
Research Article
Human Natural Regulatory T Cells Recognize Peptides of the Heavy Constant Region of IgG with High Sequence Homology with Peptides Derived from Pathogens
Jasmine Wang and Alessandra Franco*
University of California San Diego, School of Medicine, Department of Pediatrics, USA
Alessandra Franco, University of California San Diego, School of Medicine, Department of Pediatrics, 9500 Gilman drive, La Jolla CA, USA.
Received Date: June 15, 2020; Published Date: July 22, 2020
Abstract
Human natural regulatory T cells (nTreg) that control the differentiation of pro-inflammatory T cells recognize the heavy constant region of immunoglobulins G (Fc). We defined 16 peptides, 15 amino acids long, out of the whole Fc sequence immunogenic in healthy donors and patients with Rheumatoid Arthritis, a systemic autoimmune disease. In Immunology it is still debated how the thymic selection of T cells that recognize “self” peptides occurs as these T cells could be potentially harmful. However, nTreg recognize “self” and are very important in controlling the immune homeostasis. Here we show by screening the Immune epitope Data Base (IEDB) that the immunodominant Fc peptides that stimulate nTreg have a very high sequence homology with “non-self” peptides derived from a variety of pathogens. These results suggest that T cells with T cell receptors that recognize “self” peptides could be rescued from the negative selection in the thymus by engaging antigenic peptides with high sequence homology and or that pathogens may inhibit the immune regulation by jeopardizing nTreg expansion by T cell receptor antagonism.
Introduction
In Immunology it is very debated the thymic selection of T cells that recognize “self” antigens, particularly in the light of novel data suggesting that natural regulatory T cells (nTreg), essential in maintaining the immune homeostasis, recognize “self” peptide epitopes. We characterized in healthy adult donors and patients with Rheumatoid Arthritis (RA), a systemic autoimmune disease, an important specificity of nTreg that regulate the differentiation of pro-inflammatory T cells [1].
Here we explore the sequence homology of these peptides, derived from the heavy constant region (Fc) of immunoglobulin G with peptides reported in the Immune Epitope Database (IEBD). The purpose was to address a possible role for “not-self” antigenic peptides in shaping the thymic selection of nTreg and a potential mechanism for pro-inflammatory T cell epitopes with high sequence homology in inhibiting Treg expansion and exacerbating the inflammatory condition by T cell receptor (TcR) antagonism.
Method
Peptide T cell epitopes that we screened are linear sequences recognized by human T cell reported in the Immune Epitope Database (IEDB). The IEDB reports peptide epitopes derived from viruses, bacteria, fungi, tumors, allergens, and self-antigens (Table 1). The output from BLASTP contains all peptide epitopes with at least four amino acid identities, consecutive and non-consecutive, with the query Fc peptide sequence. The longest consecutive sequence homology from the output is five amino acid long. We are reporting only peptide epitopes that contain 5 or 4 consecutive amino acid overlaps with one of our 16 IgG1 Fc peptides (Tables 1).
Result
We report the wild type IgG1 Fc peptides that have five or four consecutive amino acids identical with known T cell epitopes on IEDB screened by NCBI BLAST. Three Fc peptides, Fc 21-35, Fc 61- 75, 31-45, that ranked 3rd, 7th and 10th within the immunogenicity in healthy donors and RA subjects, had a sequence homology of five consecutive amino acids with six peptide epitopes (Table 2). Four Fc peptides, Fc 181-195, 186-200, 31-45 and 276-290, that ranked 2nd, 4th, 10th and 11th within the immunogenicity in healthy donors and RA subjects had a sequence homology of four consecutive amino acids with four peptide epitopes (Table 2).
Table 1: Summary of T cell epitope categories screened for sequence homology with Fc peptides.
Table 2: Ranking of the immunogenicity of Fc peptides that expand natural regulatory T cells (nTreg) in healthy donors and Rheumatoid Arthritis Subjects.
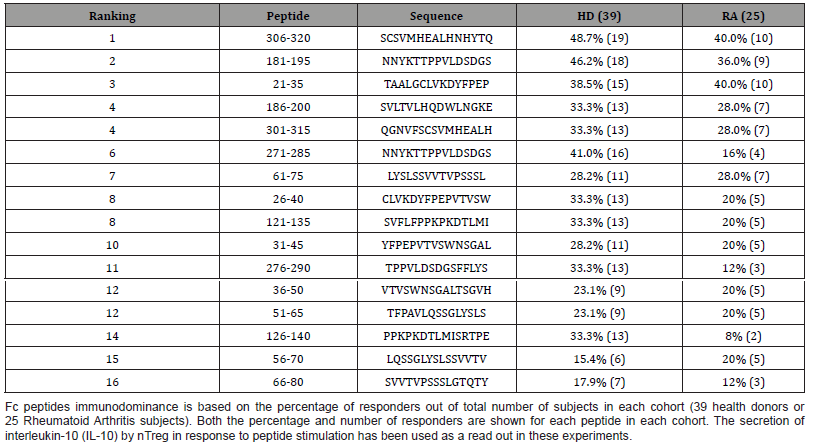
The description of the sequence homology is reported according to the ranking within the immunodominance [1] and is described in Table 2. Fc 21-35 ranked 3rd within the immunogenicity, recognized by nTreg in 38.5% in healthy donors (15 out of 39) and by nTreg in 40% RA subjects (10 out of 25). Fc 21-35 is an HLA DRB1*12:01 binder with intermediate affinity [1]. Fc 21-35 has five consecutive amino acids sequence homology, LVKDY, with mouse (Mus musculus) high molecular weight urine protein S100-A15A peptide 81-95 (Table 2, #1). The epitope was tested on mouseallergic donors [2]. Fc 21-35 also has five consecutive amino acids sequence homology, VKDYF, with the human parvovirus B19 capsid protein VP1 (P07299.1) peptide epitope 191-210 (Table 2, #2) [3].
Fc 61-75 ranked 7th in the immunogenicity, was immunogenic in 28.2% of the healthy donors (11 out of 39) and in 28% of RA subjects (7 out of 25) (Table 1). Fc 61-75 is a pan-HLA Class II binder that binds to HLA DRB1*01:01, *04:01, *04:05, *07:01, *08:02, *09:01, *15:01, DRB4*01:01, and DQB1*03:01, *06:02 [1]. Fc 61- 75 has five consecutive amino acids VVTVP identical to the human immunoglobulin binding protein BiP10 peptide 156-175 (Table 2, #3). The BiP10 peptide is a rheumatoid arthritis autoantigen that was tested in PBMC derived from RA subjects, subjects with systemic lupus erythematosus (SLE), and healthy donors [4]. The peptide epitope is an HLA-DRB1*04 binder as Fc 61-75.
Fc 61-75 shares the same five consecutive amino acids homology, VVTVP, with Timothy grass (Phleum pratense) pollen protein peptide 45-59 (Table 2, #4). The Timothy grass peptide was derived from a combined transcriptomics and proteomics analysis of the pollen protein and was identified as a novel T cell epitope predicted to be pan-HLA binder as Fc 61-75 [5].
Fc 31-45 that ranked 10th in immunogenicity, recognized by nTreg from 28.2% (11 out of 39) of healthy donors and by nTreg from 20% RA subjects (5 out of 25) (Table 1). Fc 31-45 has five consecutive amino acids homology, SWNSG, with the immunodominant peptide in Dengue Virus 2 nonstructural protein NS3 peptide 336-350 (Table 2, #5) [6]. Fc 31-45 has also a sequence homology of five consecutive amino acids, TVSWN, with SARSCoV- 2 ORF1ab polyprotein peptide 1361-1375 (Table 2, #6) [7].
Fc 181-195 is an immunodominant peptide that ranked 2nd in immunogenicity, recognized by 46.2% of the healthy donors (18 out of 39) and 36% of the RA subjects (9 out of 25) (Table 1). It is a pan-HLA Class II binder that binds to HLA DRB1*04:05, *07:01, *09:01, DRB4*01:01, and DQB1*02:01, *03:02 [1]. Fc 181-195 has four consecutive amino acids, LTVL, with Prevotella copri general secretory (Sec) signal protein peptide 2-20 (Table 2, #7), identified from T cell responses in synovial tissues and in PBMC. The peptide stimulates IFN-γ secretion (Th1 type response) in new-onset RA patients and is predicted to bind 25 HLA-DR molecules [8].
Fc 186-200 is an immunodominant peptide that ranked 4th in immunogenicity, recognized in 33.3% healthy donors (13 out of 39) and 28% RA patients (7 out of 25) (Table 1). It is a pan-HLA Class II binder that binds HLA DRB1*15:01, DRB4*01:01 and DPB1*02:01, *04:01, *04:02, *05:01 [1]. Fc 186-200 has four consecutive amino acids, HQDW, with human beta herpesvirus 6B immediateearly protein IE2 peptide 428-445 (Table 2, #8) (IEDB Reference: 1034573).
Fc 31-45 ranked 10th in immunogenicity, recognized in 28.2% (11 out of 39) healthy donors and in 20% RA subjects (5 out of 25) (Table 1). Fc 31-45 has four consecutive amino acids, WNSG, with Borrelia burgdorferi ZS7 outer surface protein A precursor peptide 204-223 (Table 2, #9) that was recognized by T cells derived from treatment-resistant Lyme Arthritis patients [9].
Fc 276-290 ranked 11th in immunogenicity, recognized in 33.3% of healthy donors (13 out of 39) and in 12% of RA patients (3 out of 25) (Table 1). Fc 276-290 has four consecutive amino acids, FFLY, with Ebola virus envelope glycoprotein precursor peptide (Table 2, #10) 152-266 [10].
Discussion
Here we report that immune modulatory peptides that stimulate human nTreg share a very high sequence homology with a variety of pathogens and allergens. Five viral peptides from Human Parvovirus, Dengue virus, SARS-CoV-2, human beta Herpes virus and Ebola virus share as much as five and four consecutive amino acid with Fc 21-31, Fc 31-45, Fc 186-200, 276-290. Two bacterial peptides derived from Prevotella copri and Borrelia Burgodofery ZS7 share four consecutive amino acids with Fc 181-195 and 31-45. Two peptides involved in allergy, mouse urine protein and Timothy grass pollen have a sequence homology of five amino acid with Fc 21-35 and 61-75 (Table 3).
Table 3: T cell epitopes with 5 and 4 consecutive amino acid sequence homologies with Fc peptides.

These results are very intriguing as they may explain the positive thymic selection of “self” specific T cells that are anticipated to be negatively selected to avoid autoimmunity. Most recently has been reported in a murine model that Treg can be rescued in the thymus from negative selection via T cell receptor (TcR) engagement with not-self peptides/MHC complexes [11]. In humans, these experiments cannot be performed because of lack of access to tissues but the high sequence homology that we found with a variety of antigens may explain the rescue of nTreg that recognize “self” epitopes. On the other hand, peptides from antigens with high sequence homology with peptides that stimulate nTreg may serve as TcR antagonist and inhibit the immune regulation [12,13].
Acknowledgement
This study has been funded by the NIH NIAID (RO1AI43586 to AF). The authors thank Dr. Barry J. Grant for helpful suggestions and Dr. Li-En Hsieh for ranking the immunodominance of the Fc peptides.
References
- Hsieh LE, J Sidney, JC Burns, DL Boyle, GS Firestein, et al. (2021) IgG Epitopes Processed and Presented by IgG (+) B Cells Induce Suppression by Human Thymic-Derived Regulatory T Cells. J Immunol 206(6): 1194-1203.
- Schulten V, L Westernberg, G Birrueta, J Sidney, S Paul, et al. (2018) Allergen and Epitope Targets of Mouse-Specific T Cell Responses in Allergy and Asthma. Front Immunol 13(9): 235.
- Kasprowicz V, A Isa, T Tolfvenstam, K Jeffery, P Bowness, et al. (2006) Tracking of peptide-specific CD4+ T-cell responses after an acute resolving viral infection: a study of parvovirus B19. J Virol 80(22): 11209-11217.
- Shoda H, K Fujio, K Sakurai, K Ishigaki, Y Nagafuchi, et al. (2015) Autoantigen BiP-Derived HLA-DR4 Epitopes Differentially Recognized by Effector and Regulatory T Cells in Rheumatoid Arthritis. Arthritis Rheumatol 67(5): 1171-1181.
- Schulten V, JA Greenbaum, M Hauser, DM McKinney, J Sidney, et al. (2013) Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc Natl Acad Sci U S A 110(9): 3459-3464.
- Rivino L, EA Kumaran, V Jovanovic, K Nadua, EW Teo, et al. (2013) Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol 87(5): 2693-2706.
- Snyder TM, RM Gittelman, M Klinger, DH May, EJ Osborne, et al. (2020) Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv.
- Pianta A, S Arvikar, K Strle, EE Drouin, Q Wang, et al. (2017) Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients With Rheumatoid Arthritis. Arthritis Rheumatol 69(5): 964-975.
- Kamradt T, B Lengl-Janssen, AF Strauss, G Bansal, AC Steere (1996) Dominant recognition of a Borrelia burgdorferi outer surface protein A peptide by T helper cells in patients with treatment-resistant Lyme arthritis. Infect Immun 64(4): 1284-1289.
- Powlson J, D Wright, A Zeltina, M Giza, M Nielsen, et al. (2019) Characterization of Antigenic MHC-Class-I-Restricted T Cell Epitopes in the Glycoprotein of Ebolavirus. Cell Rep 29(9): 2537-2545 e2533.
- Owen DL, SA Mahmud, LE Sjaastad, JB Williams, JA Spanier, et al. (2019) Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol 20(2): 195-205.
- Sette A, J Alexander, J Ruppert, K Snoke, A Franco, et al. (1994) Antigen analogs/MHC complexes as specific T cell receptor antagonists. Annu Rev Immunol 12: 413-431.
- Franco A, C Ferrari, A Sette, FV Chisari (1995) Viral mutations, TCR antagonism and escape from the immune response. Curr Opin Immunol 7(4): 524-531.
-
Jasmine Wang, Alessandra Franco. Human Natural Regulatory T Cells Recognize Peptides of the Heavy Constant Region of IgG with High Sequence Homology with Peptides Derived from Pathogens. Arch Biomed Eng & Biotechnol. 5(4): 2021. ABEB.MS.ID.000618.
-
Homology, Peptide epitopes, Antigens, Musculus, Immunogenicity, Urine protein, Rheumatoid arthritis
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






