Opinion
Opinion
Efficacy of Weka for Medical Data Mining: Ambulatory Blood Pressure Monitoring as A Case-Study
Chuiko GP1*, Darnapuk YS2, Dvormik OV3, Honcharov DA4 and Yaremchuk OM5
1Professor, Department Computer Engineering, Ukraine
2Senior Lecturer, Department Computer Engineering, Ukraine
3Associate Professor, Department Computer Engineering, Ukraine
4Head of Lab, Information Computer Center, Ukraine
5Professor, Dean of Medical Institute, Ukraine
Chuiko G P, professor, Department Computer Engineering, Ukraine.
Received Date: April 24, 2023; Published Date: May 11, 2023
Introduction
Ambulatory blood pressure monitoring (ABPM) gradually gained weight from the early 1960s. ABPM, alias 24-hour blood pressure trial, is one routine medical test for diagnosing and prophylactics of circulation upsets, particularly hypertension, even disguised [1]. Many medical databases exist concerning ABPM, sometimes quite detailed and ponderous enough, like [2]. Medical databases can comprise dozens and hundreds of various attributes, hundreds and even thousands of instances. Detalization makes such datasets rich on the one hand but just “big data” on the other. “Big data” scares clinicians, who are primarily unfamiliar with modern data mining means. One innovative solution is comprehensive visualization and graphical user interface (GUI) for medical data mining tools.
WEKA is modern, Java-based software for data mining, successfully developing the GUI methods and visualizing means during the last two and half decades [3]. Despite WEKA having an evident power for practical data mining, this software is not too popular in Ukraine; We know only one short course dedicated to WEKA that is reading in Igor Sikorsky Kyiv Polytechnic starting in 2018 [4]. The authors hope this report could help to well publicity for outstanding soft, which is free besides.
Let us consider the initial ABPM dataset [2]. The study was performed by 159 women and 101 men aged between 14 and 92. So the dataset contains 270 instances. The ABPM records were attained by a qualified cardiologist from the Serif Medical Teaching Hospital, Cardiology Department, in Algeria [2]. Each patient was characterized by 40 attributes and six levels (circadian rhythm manifesting, blood pressure load, morning surge, validity, pulse pressure, and blood pressure variability). Thus, it looks like genuine “big data” from the point of view of a clinician.
Let us preprocess the initial dataset with WEKA. One can remove all the database’s invalid instances (exemplars) from the beginning. Besides, one can remove the labels “Validity,” “Blood Pressure Variability,” and attribute “Interrupt” because the two first are inherited in all remained instances, and this third contrarily is now absent there in the whole. As a result, instead of six levels (possible classes) of Table 1, there left only four: Circadian Rhythm, Pulse Pressure, Blood Pressure Load, and Morning Surge. Thus, one has a reduced but fully valid data set with 36 attributes (including four possible binary classes) and 185 instances. The dataset is still bulky enough, but “the process already started.”
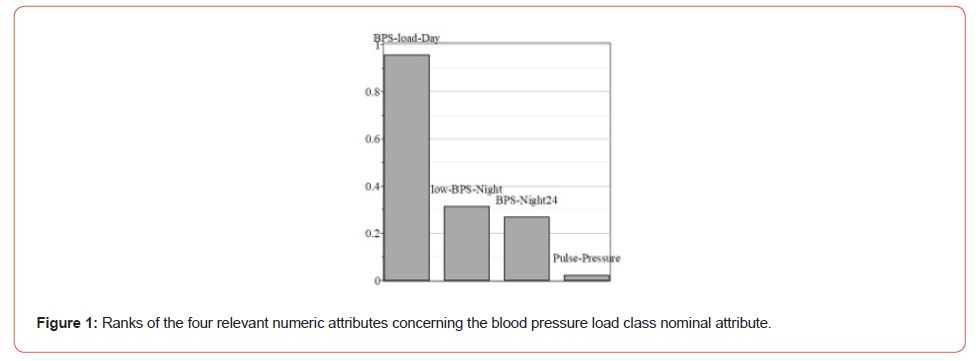
Blood pressure load is the percentage of blood pressure readings equal to or above the cutoff point for determining elevated readings. Thus, blood pressure load provides valuable information for diagnosing hypertension. Higher this parameter, so more likely it is hypertension for a patient. There are two unequal and imbalanced classes of the patient in our dataset: 124 with blood pressure load and 61 without one. Let us apply the attribute selection filter to this dataset. Weks has several similar filters, and Cfs Subset Eval is one of them. It evaluates the worth of a subset of attributes by considering each feature’s predictive ability and the degree of redundancy between them. Subsets of attributes highly correlated with the class while having low intercorrelation are preferred. Only four numeric attributes from 36 were overcome throughout this filter. These are Pulse pressure (Pulse-Pressure), Night Blood pressure Systolic Average (BPS-Night24), Day Blood pressure Systolic load value (BPS-load-Day), and the lowest Blood pressure Systolic night (low-BPS-Night).
WEKA allows the ranking of obtained attribute subsets. (Figure 1) shows the relative ranks of these attributes. The rating was obtained using the Info Gain Attribute Eval algorithm that evaluates the worth of an attribute by measuring the information gained concerning the class.
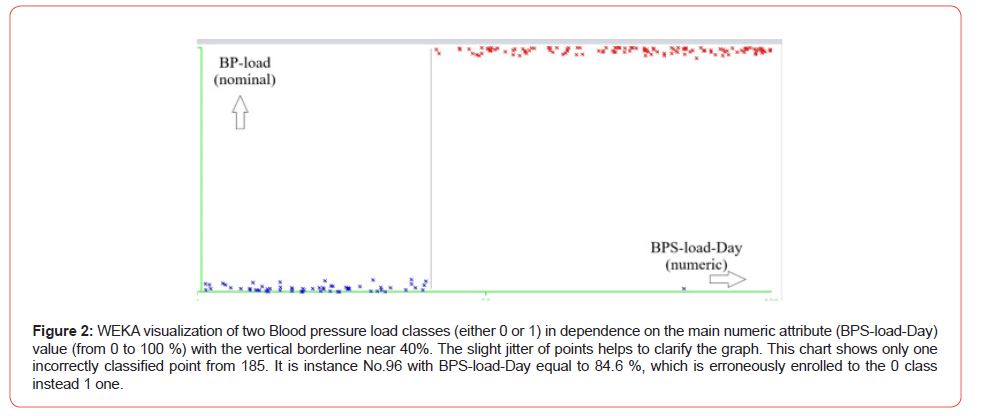
The attribute is BPS-load-Day, or the day Systolic blood pressure load value (from 0 to 100%) has the highest rank, the most correlated with the nominal classes. Hence, the visual classification is most reasonable by ty this attribute (Figure 2). Indeed, Figure 2 correctly classified 184 instances from 185. Thus, the typical problems of Machine Learning and data mining, such as classification and selection of attributes and their ranking, are well-solvable within WEKA software even without expertise in Java. Mainly it is due to the detailed visualization of all of the mining operations within WEK a, and especially it concerns the last versions of the software. The authors, among them, is the dean of the Medical Institute of our university, are convinced of the necessity of WEKA courses in the education process. It touches both kinds of students: computer engineers and medics, though the content of these courses should be different, to be sure.
Acknowledgment
None.
Conflict of Interest
No conflict of interest.
References
- (2021) https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds).
- Patterson D, Slack J (1972) Lipid abnormalities in male and female survivors of myocardial infarction and their first-degree relatives. Lancet 1(7747): 393-399.
- Epstein F (1976) Genetics of ischemic heart disease. Postgraduate Medical Journal 52(610): 477-480.
- Miyazawa K, Ito K (2021) Genetic Analysis for Coronary Artery Disease Toward Diverse Populations. Front. Genet 12: 766485.
- Kessler T, Schunkert H (2021) CAD Genetics Enlightened by GWASs. JACC: Basic to Translational Science 6(7): 610-623.
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, et al. (2002) Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nature Genetics December 32(4): 650-654.
- Koyama S, Ito K, Terao C, Akiyama M, Horikoshi M, et al. (2020) Population-specific and Trans-ancestry Genome-wide Analyses Identify Distinct and Shared Genetic Risk Loci for Coronary Artery Disease. Nat Genet 52: 1169-1177.
- Roberts R (2014) Genetics of Coronary Artery Disease: An Update. Methodist Debakey Cardiovasc J 10(1): 7-12.
- Sioziou A, Katifelis H, Legaki E, Patelis N, Athanasiadis D, et al. (2018) Expression of miR21, miR122, miR146a and miR196 in Symptomatic Carotid Disease. Int Cardiovasc Res J 12(1): 7-12.
-
Chuiko GP*, Darnapuk YS, Dvormik OV, Honcharov DA and Yaremchuk OM. Efficacy of Weka for Medical Data Mining: Ambulatory Blood Pressure Monitoring as A Case-Study. On J Cardio Res & Rep. 7(3): 2023. OJCRR.MS.ID.000661.
-
Circadian rhythm manifesting, Blood pressure load, Morning surge, Validity, Pulse pressure, Blood pressure variability, Hypertension, Cardiology, Circulation upsets
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.