 Research Article
Research Article
Inhibition of BRD4 Attenuates Inflammation and Oxidative Stress in Retina
Chih-Hung Chen1,2, SiDe Li1,2, Elena Rusinova1, Guangtao Zhang1, Ming-Ming Zhou1, Martin J Walsh1,2,3 and Joseph W Eichenbaum4*
1Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, USA
2Departments of Pediatrics, Icahn School of Medicine at Mount Sinai, USA
3Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, USA
4Department of Ophthalmology, Icahn School of Medicine at Mount Sinai, USA
Joseph W Eichenbaum, Department of Ophthalmology, Icahn School of Medicine at Mount Sinai, New York, USA.
Received Date: July 04, 2022; Published Date: July 29, 2022
Abstract
Aim: To determine how oxidative stress in the zebrafish retina alters the expression of genes linked with stress pathways and how different
inhibitors compare in their effectiveness in suppressing damage linked with specific cellular processes.
Methods: To explore these issues, we applied a higher level of Rose Bengal and cold white light, HRBCWL, to the zebrafish eye, to mimic the
onset of macula degeneration (AMD). Then, either intravitreally injected BET bromodomain inhibitor, (BRDi), MS255, or VEGF inhibitor, (VEGFi),
Ranibizumab, approved for clinically treating AMD, were used, and compared for their respective retinal histologic and transcriptional outcomes.
Key Findings: Examination, after HRBCWL treatment, revealed greater retinal layer height elevation after VEGFi than BRDi and the transcript
patterns of the two treatments were strikingly different. After the BRDi there was loss of a core inflammatory gene signature and a moderate
decrease in Nrf2 and Keap1, (oxidative stress gene) expression. However, VEGFi treatment did not affect Nrf2 or Keap1 but showed reduction of
mitochondrial genes Noxa1 and Ucp3 activity. BRDi suppressed the expression of Sod2 and Sirt2. VEGFi had no effect on their expression. Sirt1
expression showed elevation after HRBCWL followed by BRDi, but mild reduction after VEGFi. MULAN, a key mitochondrial protein involved with
NFκΒ signaling, was also reduced after BRDI.
Significance: This study illustrated the first potential and differentiating role for BRDI in mitigating oxidative stress/retinal degeneration
transcription factors (Nrf2 and Mul1b) and that the BRDi’s genomic signature is distinct from VEGFi. This could imply that management of AMD
could be varied in targeting different transcripts and signaling pathways either individually or in combination as a therapeutic approach for AMD.
Keywords: Bromodomain inhibitor; VEGF inhibitor; Retinal degeneration; Transcription; Oxidative stress; Autophagy; Nrf2; Mul1b; Mitochondria
Introduction
As the U.S. population ages, macula degeneration, which reduces central vision, is the leading cause of vision loss in the over 65 age group. Unfortunately, there are few therapeutic options available for the treatment and relief of disease progression for these patients. While oxidative stress is believed to play a crucial role in the development and progression of macula degeneration, little is understood regarding the early behavior of transcriptional pathways that regulate the response to oxidative stress in the retina. Recent studies have suggested the multidimensional aspects of retinal epithelial cell biology that contribute to the anti-oxidative stress response, inflammation, mitochondria damage, autophagy, hypoxia, unfolded protein stress response all which coalesce around the epigenetic events leading to the development of macula degeneration (AMD) [1,2]. However, the specific transcriptional mechanisms and genetic networks remain poorly understood. Oxidative stress, including an increase in the presence of inflammatory cells, has been shown to be associated with an increase of “drusen-related” proteins in dry AMD [3]. While AMD is linked by susceptibility through genetic variation and polymorphisms in genes which include: (1) Complement Factor H, CFH, [4,5], in dry and wet AMD, (2) Htra1/ARMS 2, [6], (3) Hepatic Lipase c, [7], and (4) TIMPS, a metalloproteinase, [8] in Sorbsby’s macula degeneration, an early form of AMD, [9,10] there has not been early definitive genomic data in the underlying etiology of AMD [1]. While such genes are implicated and seem to support the intersection between the multiple pathways linked through an oxidative stress response a more detailed investigation to understand the underlying mechanisms for their association remains untested. The correlation of these genes to prognostic outcomes, after the use of e.g., a VEGFI for wet AMD treatment, show that in patients with specific ARMS2 alleles, there was little correlation with improvement to visual impairment [11]. On the other hand, regarding validation of a prediction for the progression to advanced macula degeneration, Seddon et. al found increased risk with the ARMS2 variant allele for progression to neo-vascular NV AMD [12]. Also, drusen and phenotypic changes in association with genetic alteration suggests the progression of AMD [13]. Added to that, genetic changes associated with key risk factors including age, sex, education, body mass index, smoking, and baseline AMD status may have AMD predictive value as well [13]. This is especially true in those carrying the CFH alleles where the odds of AMD were 3.3 times greater among women with low lifestyle score and two high risk CFH alleles compared to those women who had very healthy lifestyle and two low risk CFH alleles [4,14- 17]. With similar consideration, the odds of AMD were highest in those with deficient vitamin D and 2 risk alleles for the CFH and CFI genotypes, suggesting a synergistic effect between vitamin D status and genetic predisposition [14-17]. Of note, individuals with high CFH and no ARMS2 risk alleles and taking the AREDS, age related eye disease antioxidants formulation, had increased progression to NV, neovascularization, compared with placebo [18]. Although an underlying genetic predisposition to AMD is strongly suggested, an increasing emphasis has been placed on the contributing epigenetic parameters that regulate the response to oxidative stress. These epigenetic modifiers may be active in inflammation [19-21,] changes in cellular metabolism generating glycation products [22,23], protein misfolding, autophagy [24], and mitochondrial dysfunction [25] in the genesis of AMD. A dynamic interplay of these elements with the overall architecture of the genomic transcriptome could account for the complexities of their interactions. At the center of this process is where nuclear factor kappaB (NFκΒ or RelA) directs key pathways evoking inflammation, cell damage, and apoptotsis [26]. The underlying inflection point places Nrf2, as a transcription factor, at the core [27] of the cyto-protective function in Retinal Pigment Epithelium, [25,28,29] as well activating an antioxidant transcriptional response through Superoxide dismutase (SOD1), Hemoxygenase2 (Hmox2), and Catalase (Catl) stimulated after oxidative stress in balancing transcription factor action [ 15,29]. This interpretation may suggest a dynamic interplay between stress and anti-stress modulating factors mediated at the transcriptional levels. Such information has provided the rationale of downmodulating the damaging transcriptional responses by re- balancing the oxidative stress responses, inflammatory signals, and signals leading toward cellular damage signaling such as apoptosis [25]. Here, we examined the transcriptional outcomes after oxidative stress, the key regulatory networks at play, and the role of chromatin modifiers in the reduction of acute retinal degeneration. Examining the phenotypic changes in a wellcharacterized and highly reproducible retinal oxidative stress model [30] we tested the efficacy of two epigenetic inhibitors of the BET class of transcriptional activators [31] introduced after high retinal oxidative stress. We learned that MS55 could attenuate the advances of phenotypic retinal degeneration at least as well as VEGF inhibitor, ranibizumab. We also noted that the anti-oxidative response by MS255 corresponded with an elevation of antioxidative stress Nrf2 genes and that this corresponded with genes linked to mitochondrial proteosomal function, Sgk1 and Mul1b. We now provide a fundamental understanding for the transcriptional network architecture that may underlie oxidative stress responses in AMD.
Materials and Methods
Retina Preparation and Histology
Wild-type adult, over 1 year old zebrafish, were purchased from That Fish Place, (Lancaster, PA). The zebrafish were kept in a receiving stock tank for 1 month at a 12-hour light and dark cycle and reared at 28°C according to standard procedures. All procedures, anesthesia, Rose Bengal preparation and use, cold white light and its metering described here were approved by the ISMMS Institutional Animal Care and Use Committee (IACUC) as previously described [30].
Anesthesia
1 or 2 cc of tricaine methanesulfonate, concentration 164mg/ ml was placed in a 150cc zebrafish tank water sponge based small rectangular swim container. One zebrafish at a time was netted from the larger tank and placed in the swim container until anesthesia took which was usually about a minute or less.
Rose Bengal
The right eye consistently underwent intravitreal injection of Rose Bengal and BRDI or VEGFI. Rose glo sterile ophthalmic strip, (Rose Stone Enterprises, Alta Loma, CA) were used to prepare 12mg/ ml Rose Bengal in distilled water. Microliter #62-gauge (Hamilton Co., Reno NV) syringe and needle, 33GARN-37 30 DEG, were used to inject 1uL of Rose Bengal (and BRDI or VEGFI) intravitreally under microscopic control (Mentor Microscope, CMIII, Randolph, Mass). The injection was on the caudal side of the globe, just behind the ciliary body near the 9 o’clock limbus.
Fiberoptic Cold White Light, Light Meter
Gossen Mavolux 5032C light meter (Germany) was used to measure the lumens in lux of the Volpi Intralux 5000 cold white coherent fiber optic light (120-volt, 185-watt, 50/60 Hz, (Switzerland). After confirming the light energy with the light meter, the fiber optic cable was placed into the water bath over the zebrafish cornea. The time of the lesion induction from removal of the fish from the anesthesia container to completion of the luminance was under 5 minutes. Fish breathing and heartbeat were monitored through the microscope. All cold white light exposures were for 1 minute at 103x103 lux. Controls were immediately transferred to a post operative tank where they swam for two hours under usual light conditions and were then euthanized with standard tricaine protocol, 0.4 mg /ml tricaine solution.
Experimental Groups
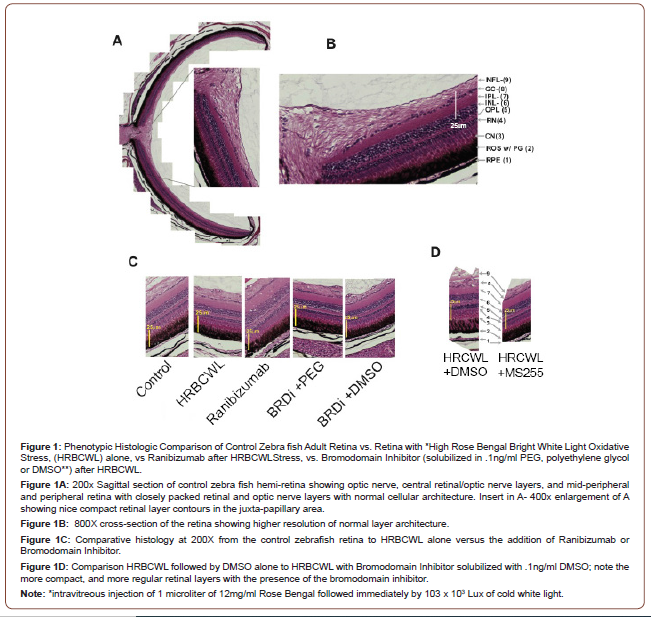
BRDI fish were injected intravitreally after cold white light exposure with 1uL of 0.01ng/ml MS255 [31]. VEGFI delineated fish were injected intravitreally after cold white light exposure with 1uL of .01mg/ml Ranibizumab. After experimental BRDI and VEGFI fish had their injections, they were placed in separate tanks for a 2-hour swim and then euthanized for retina removal. Fish selected for three-day furlough analysis were replaced in their respective BRDI and VEGFI experimental tanks and removed after three days and euthanized according to the standard procedures. The retinae were carefully removed with the retinal pigment epithelial layer attached. After the zebrafish retinae were surgically removed with 0.12 Castroviejo Forceps and Van Ness scissors under Unitron dissecting microscopic control, some of the control and experimental retinae were snap frozen in liquid nitrogen and stored at -80°C for later experiments. Other control and experimental retinae were placed in modified Davidson’s fixative, according to the instructions of (Excalibur Pathology Inc, Norman, Oklahoma) who performed the Hematoxylin and Eosin slide preparation. The retinal histologic sections of experimental and control retinae were then photographed. The retinal layers were then graded for comparative height and cell width, and retinal layer architecture, such as the presence of vacuoles or height and width of the retinal pigment epithelial layer, the nuclei of the cones, the nuclei of the rods, the plexiform layers, and the ganglion cell layer. Measurements of the retinal layers in each of the control and experimental groups were made using the Olympus DP70 camera software. Measurements were made for each retinal layer and indicated in Figure 1.
RNA Extraction, RNA Sequencing and Quantitative PCR (qPCR)
As indicated above, all zebrafish retinae were surgically dissected and immediately immersed in RNA ready solution in preparation of RNA extraction and isolation. All RNA Isolations were performed using the standard Trizol protocol. Reverse transcription was accomplished using the Superscript III kit from Invitrogen with 2ug total RNA. RT-PCR reactions were prepared with 50ng cDNA per reaction and RedTaq reverse transcriptase mix (Sigma) and amplified for 45 cycles. Annealing temperature was 58 degrees C and extension time was 3 minutes. Samples were then run on 0.75% weight to volume (w/v) agarose with 0.05% w/v ethidium bromide for ultraviolet light (UV) detection. RT-qPCR reactions were prepared with 20ng cDNA per sample and set up according to the Promega GoTaq 2-Step Sybr Green kit using a fast 2-step protocol on the Agilent Mx3000P qPCR system and initial melting time of 3 minutes. For short read sequencing, total RNA was isolated from sample sets of approximately 20 retinae using standard TRIzol® (phenol/chloroform) isolation procedures. RNA quality was assessed by an Aglient bioanalyzer. RNAs with a RIN score of 9.0 or greater were used to generate cDNA libraries following treatment with the Ribo-Zero rRNA removal kit (Illumina). RNA-Seq library preparation was performed at the Weill Cornell Medical College Genomic Core facility (New York) using the TrueSeq RNA sample preparation kit (Illumina RS-122-2001) as per manufacturer’s recommendations. Samples were sequenced by the Illumina HiSeq 2500 platform (Illumina HiSeq 2500) as 100 bp pair-ended reads.
RNA Sequencing
RNA-seq reads were aligned to the GRCz11 Danio rerio build annotation for the zebrafish genome and quantified using the Tuxedo Suite (Bowtie, Tophat, Cufflinks) tools [32-34]. Contaminant (aligned) RNA-Seq reads was filtered and aligned to several mouse reference databases including the zebrafish genome (GRCz11, NCBI Build 37), RefSeq exons and splicing junctions using BWA alignment algorithm [35]. The reads that were uniquely aligned to the exon and splicing-junction sites for each transcript were then counted as expression level for a corresponding transcript and were subjected to log2 transformation and global median normalization. Differentially expressed genes were identified by the R package DEGseq [36] using a false discovery rate (FDR) < 0.001 and foldchange >1.5.
Quantitative PCR Data Analysis
Gene expression specific primers used for this study are listed in Table 1. Hprt RNA was assayed as loading control. Dissociation curves were also obtained. All results for each technical replicate are normalized to the relevant control samples; therefore, controls show no error and standard error bars shown report deviation of all normalized replicates from their respective controls. All experiments reported are triplicate (n=3) experiments.
Table 1: Danio Rerio Primer List.

Results
Transcriptome–Wide Alterations Define Key Gene Transcript Signatures that Embody Both Oxidative and Inflammatory Stress Pathways
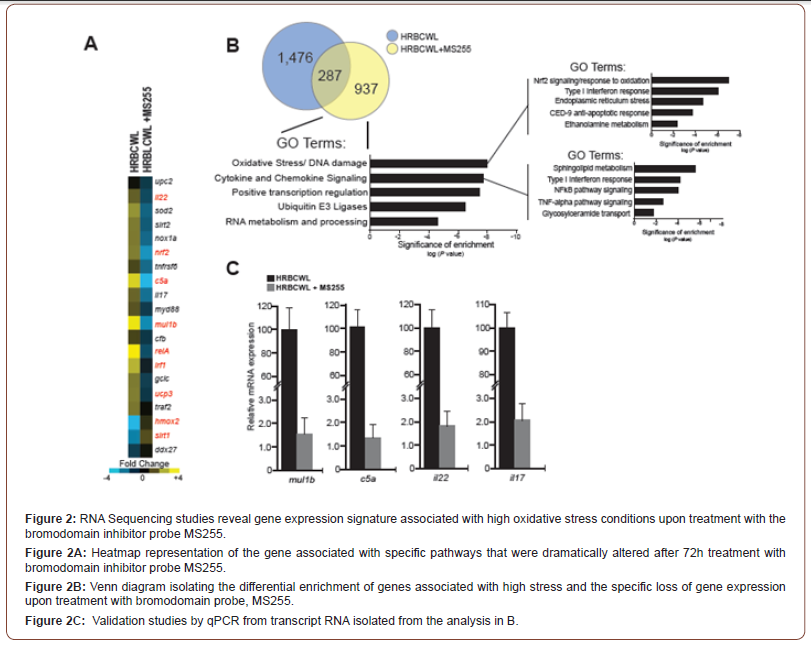
Three days after adult zebra fish sustained hi level retinal oxidative stress, (intra vitreous injection of 1uL of 12mg/ml Rose Bengal followed by 103 x 103 lux cold white light), we observed that the use of BRDI MS255 immediately after the photochemical stress, in a more pronounced fashion than VEGFI, Ranibizumab, helped attenuate the stress pattern of retinal layer height elevation and cell enlargement. This retinal oxidative stress pattern was noted in this paper and previous work, [30]; see Figures 1C and Figure 1D: control, oxidative stress, oxidative stress followed VEGFI, oxidative stress followed by 2 distinct BRDI, oxidative stress followed by 1 lipid solvents sans BRDI. We evaluated transcriptome wide alterations that were related to the BRDI. Nrf2, a transcription factor that is activated in the cytoplasm by oxidative stress and is located in trans to the nucleus to activate Antioxidant Releasing Elements (ARE) -containing genes, such as Sod2 and Hmox1, and other strong antioxidants, was elevated after oxidative stress but reduced after oxidative stress that was immediately followed by either of the anti-inflammatory BRDI, (see Figure 2). In a similar fashion mTOR, SGK1, (not shown in Figure 2), transcription factors related to metabolic control of glucose metabolism/ glycolysis pathways were also up-regulated after oxidative stress and downregulated with BRDI epigenetic modifiers. C5a, il22, and il17, inflammatory mediators, which may be linked to NFKb (RelA), also showed gene upregulation after oxidative stress that was followed by down-regulation after either of the BRDI. Figure 2C Mul1b, the zebrafish gene that produces an E3 ubiquitin ligase and targets proteins for ubiquitination and helps regulate mitochondrial dynamics, and mitophagy, had its expression up regulated after oxidative stress and reduced after treatment with the BRDI MS255, (Figure 2C). Three days after adult zebrafish sustained high level retinal oxidative stress, (intra vitreous injection of 1uL of 12mg/ml Rose Bengal followed by 103 x 103 lux cold light), we observed that the use of two distinct BRDI immediately after the stress, in a more pronounced fashion than VEGFI resulted in less retinal layer height elevation, helped attenuate the stress pattern of photoreceptors, their nuclei and pigment epithelial cell enlargement. This pattern in the stressed zebrafish retina may be analogous to the retinal layer thickening and elevation seen on optical coherent tomography in macula degeneration and diabetic macula edema.
Phenotypic Analysis of the Normal Retina Versus Treatment with VEGF Inhibition or by Using a Bromodomain Inhibitor
In Figure 1A, we show normal histology of a sagittal section of the adult zebrafish retina within an exploded panel highlights normal histology at a higher resolution. Figure 1B shows normal layered architecture at higher microscopic resolution with normal retinal cell height and thickness. Indicated are the specific retinal layers. Figure 1C, shows that HRBCWL has the elongation and enlargement of the cone nuclei, larger vacuoles in the inner nuclear layer and nerve fiber layer. Moreover, Figure 1C shows that the phenotypic pattern after VEGF inhibitor (Ranibizumab) or bromodomain inhibitor (MS255) treatments are histologically distinct, However, the presence of DMSO or PEG with MS255 shows a similar histological pattern. We note the similarity of the retinal layers with each of the inhibitors as compared to the enlarged retinal cell layers with HRBCWL alone in three independent histological photomicrograph comparisons. Even with exposure to the retino-toxic reagents of polyethylene glycol (PEG) or dimethyl sulfoxide (DMSO) to solubilize the bromodomain inhibitor MS255, the bromodomain inhibitor results in less retinal layer elongation and enlargement than with high stress and DMSO (see Figure 1D). In Figure 1D, HRBCWL with DMSO alone is compared with HRBCWL with MS255 in the presence of DMSO. In Figure 1D note the compactness of the retinal architecture with the presence of MS255, whereas, the DMSO panel has more RPE and rod outer segment elongation, cone nuclei disarray and prominent vacuoles. This HRBCWL retinal architectural effect was like that observed previously [30]. In summary, we conclude that use of MS255, as a bromodomain inhibitor, provides a more pronounced phenotypic histological effect than the VEGF inhibitor (Ranibizumab), with more regular histological retinal layer height preservation. At an extremely low dose (0.1ng/ml) retino-toxic PEG or DMSO was used in conjunction with the bromodomain inhibitor for solubilizing the bromodomain inhibitor. It could be reasoned that the toxic agents PEG or DMSO are responsible for the preservation of retinal architecture. However, in Figure 1D we note the larger disorganization in the retinal architecture with DMSO alone after HRBCWL. Thus, the transcript expression pattern of Nrf2, Mul1b, C5a, il22 and il17 of elevation with oxidative stress and reduction with BRDI immediately after the stress illustrates a dynamic genetic signature that suggests the potential for genomic modification early in the acute stress episode.
Validation of RNA Signatures Define the Pathway Responses to Ranibizumab and the Bromodomain Inhibitor, MS255
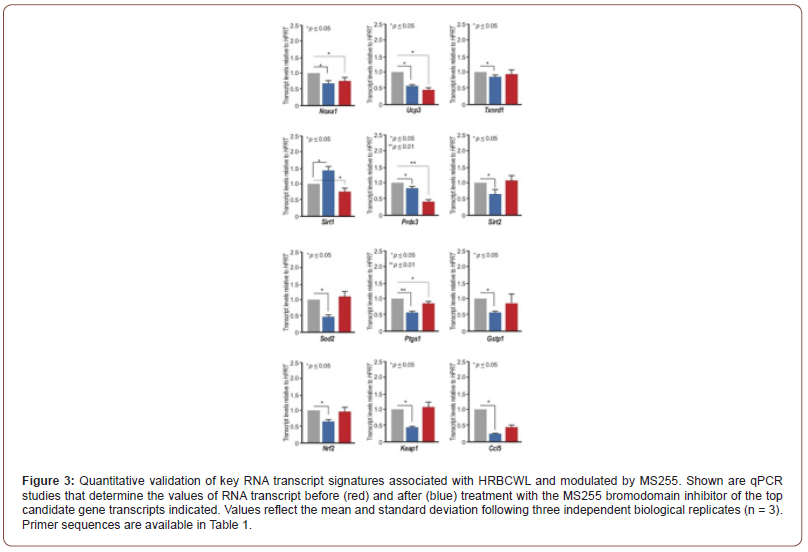
As per our studies using transcriptome-wide analysis, we took a pathway approach based on Figure 2B to illuminate the key pathways and the genes involved in the responses to either Ranibizumab or the bromodomain inhibitor, MS255 in adult zebrafish retina following HRBCWL. Our results now define the oxidative stress and inflammatory stress pathways as those pathways most directly modulated by the VEGF inhibitor (VEGFI), Ranibizumab, and the bromodomain inhibitor, MS255. We tested 12 specific genes linked to either inflammation/cytokine gene modulation or oxidative stress pathways Figure 3. These include, Nrf2, Keap1, Noxa1, and the complement protein Ccl5 Figure 3. The consequences that emerged from the treatment and validation of the transcriptome wide analysis indicate a strong and direct influence by the bromodomain inhibitor, MS255 to suppress the oxidative stress response whereas, the VEGFI, Ranibizumab early on seems to only target fewer oxidative stress pathway genes Figure 3. This would indicate that perhaps the broader effects of the bromodomain inhibitor are likely to have a more significant impact on early acute oxidative stress and inflammation transcription than VEGFI (Figure 3).
Cellular and Mitochondrial Superoxide Abundance Upon Stress is Mitigated by Administration of the Bromodomain Inhibitor MS255
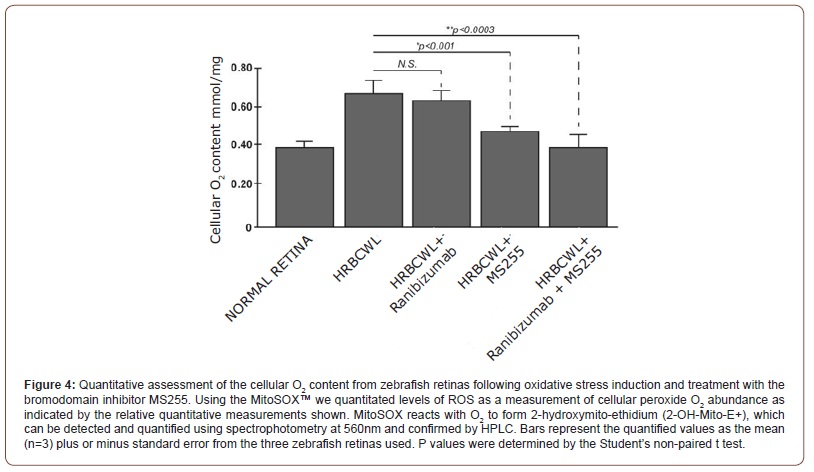
Following the treatment with Rose Bengal and intense cold white light of 103x103 lux results in the accumulation of O2 as identified as a hydrogen peroxide (H2O2) species. In this regard we measured cellular O2 content by using DHE and Mito SOX staining [37]. The production of superoxide by mitochondria can be quantified using the Mito SOX™ Red reagent. Mito SOX™ Red reagent permeates live cells where it selectively targets mitochondria. It is rapidly oxidized by superoxide but not by other reactive oxygen species (ROS) and reactive nitrogen species (RNS). The oxidized product is highly fluorescent upon binding to nucleic acid which is then quantified by spectrophotometry. Samples were taken following the preparation of retina, as described above, and measured accordingly, Figure 4. Results of our study indicate that the absorption at 572nm for fluorescent intensity of Mito SOX following PQ exposure measured by spectrophotometry is detected within normal and untreated retina sample and significant notable increases are measured upon stress induction, Figure 4. When the retinas are pretreated with either the VEGF inhibitor (Ranibizumab) or with the bromodomain inhibitor MS255, the treatment with MS255 fared better in reducing overall cellular O2 content as measured by absorption @572nm. It is also evident that when both Ranibizumab and MS255 are combined there is an added benefit, with less fluorescence from less induced O2 cellular content, Figure 4. These results would postulate that MS255 is targeting elevated O2, independently of the VEGFI pathway, whereby additive benefit of the treatments is noted (Figure 4).
Discussion
After significant acute retinal oxidative stress, in our zebrafish retina model [30], Nrf2, the transcription factor for nuclear signaling of anti-oxidative enzymes elaboration was among the statistically up-regulated (p<0.01) response with an inversely regulated Mul1b, complement C5a, interleukin 22 (il22), and interleukin 17 (il17) gene responses Figure 1. These responses may be seen as strong individual reactions to oxidative stress, or they may tie together with energy balancing pathways between the bio-energetic mitochondrial responses, the oxidative stress machinery of the cytoplasm and the transcription factors needed for oxidative stress compensation of the nucleus, which further contributes to an additional inflammatory response mediated through factors such as NFkB [27]. Nrf2 transcription factor, which has multifactorial catalytic ability, has an Neh1 domain, which allows it to hetero-dimerize with small Maf proteins. Nrf2 also has a Neh2 domain, which allows it to be bound and regulated by its cytoplasmic repressor Keap 1 [27]. Alternatively, a Neh3 domain can activate the Nrf2 transcriptional apparatus. But the Neh4 and 5 domains can bind Nrf2 to cAMP Response Element Binding Protein, CREB, which possesses histone acetyltransferase activity and can activate several nuclear anti-oxidative enzymes as well as interact with the Bromodomain proteins and initiate additional DNA/RNA transcription. An additional Neh6 Nrf2 domain may bind with proteins that will degrade Nrf2 when in stressed cells the Nrf2 protein life is protracted [27]. The BET class of bromodomain transcriptional co-activators are directly associated with chromatin by interacting directly with amino terminal acetyllysines of nucleosomal histones [31]. Among the genes that were most affected by bromodomain inhibition (to acetyllysine recognition by the BET class of bromodomains) was Mul1b (or MULAN). After high oxidative stress, Mul1b was also elevated, but was significantly reduced by the BRDI, MS255. Mul1b, (a little studied E3 ligase), operates in conjunction with E1, ubiquitin activating enzyme and E2, ubiquitin conjugating enzyme, and with ATP. [38]. E3 ligases target ubiquitination of specific proteins destined for proteosomal localization. Cullins, a family of evolutionarily conserved proteins, assemble by far the largest number of E3 ligase proteins [39,40]. Cullins 3, CUL3, binds to E3 ligase and Keap1, a negative regulator of Nrf2 and may promote Nrf2 ubiquitination by CUL3-ROC1ligase, which is a prelude for Nrf2 proteasome degradation. Blocking Nrf2 protein degradation by knocking down CUL3 can result in greater Nrf2 cytoplasmic accumulation. Alternatively, by using MS255, the levels of the Mul1b E3 ligases may have been reduced resulting in elevation of Nrf2 with greater SOD2 abundance and anti-oxidative stress gene activation. It is possible that the Nrf2- Keap cytoplasmic interaction, activated by oxidative stress, and the Nrf2E3 ligase Cul3 nuclear pathway to Nrf2 transcription factor activation for proteasome ubiquitination, are in close signal communication to mitochondrial energy sources. Mul1b is a mitochondrial protein [38], specifically oriented in a manner so that the E3 active C terminal Ring Finger is exposed to the cytosol, where it has access to the ubiquitin ligases. An intact Ring Finger (Rnf) and the correct subcellular localization are required for mitochondrial dynamics [41]. Thus, Mul1b downstream targets are likely tied to mitochondrial bioenergetics as well as E3 ligases regulating ubiquitination. Mulan has also been identified as an activator of nuclear factor kappaB (NFκΒ), referred to as RelA, which would link the mitochondria to not only nuclear signaling [42], but also the initiation of immune/inflammatory proteins, such as the complement factor H (CFH), il22 and il17 expression. Our quantitative validations of the transcriptome-wide analysis using qPCR identified the complement protein gene C5a, interleukin 22 (il22), and interleukin 17 (il17) to be among the most highly expressed after high oxidative stress, (as was the case with Nrf2 and Mul1b) but all were reduced after the use of the bromodomain inhibitor, MS255. These genes clearly correspond with stress, but it is important to note that not all inflammatory- and oxidative stress-related genes were affected by HRBCWL. This would suggest the effects are modulated possibly through specific “gatekeeper” genes. The epidemiologic papers linking lifestyle factors such as diet, exercise and smoking as well as vitamin D levels with AMD severity in association with the CFH allele presence in man [43] highlight the significance of epigenetic variables in the advance of AMD disease process. This might lead to examination of thesepotential role of epigenetic modifiers. By targeting bromodomain readers of the acetyl lysines, perhaps the harmful effects of lifestyle and high oxidative stress resulting in the chronic inflammatory and degenerative changes of AMD could be attenuated. In conclusion, our work illustrating the possible role of BRD inhibition (BRDI) with MS 255 can function either at the cytoplasmic Nrf2-Keap1/ Anti-oxidative stress pathway, or the mul1b-Nrf2E3ligaseCul3, ubiquitination pathway, or the Nrf2-Mul1b-Nfkb network in single or combination pathways after oxidative stress. This may provide insight into developing a brake on the evolution of AMD. If BRDI can serve as a brake on excess oxidative stress outcomes in cytoplasmic, nucleus, and mitochondrial energy demands, perhaps the bromodomain inhibition is expected to be beneficial over time in attenuating the binge damaging effects of oxidative stress in the evolution of adult-onset macula degeneration (AMD) and other retinal degenerative diseases.
Conclusion
In conclusion, our work illustrating the possible role of BRD inhibition (BRDI) with MS 255 can function either at the cytoplasmic Nrf2-Keap1/Anti-oxidative stress pathway, or the mul1b- Nrf2E3ligaseCul3, ubiquitination pathway, or the Nrf2-Mul1b-Nfkb network in single or combination pathways after oxidative stress may provide insight into a brake on the evolution of AMD. If BRDI 255 can serve as a brake on excess oxidative stress outcomes in cytoplasmic, nucleus, and mitochondrial energy demands, perhaps the bromodomain inhibition is expected to be beneficial over time in attenuating the binge damaging effects of oxidative stress in the evolution of adult-onset macula degeneration (AMD) and other retinal degenerative diseases.
Supplemental Information
Supplemental information includes a supplemental table for all primer sequences used for polymerase chain reaction experiments.
Acknowledgement
We thank Dr. A. Alonso of the Weill-Cornell College of Medicine’s Epigenomic Sequencing Core for advice and support for genomic sequencing and library preparation for RNA-Seq. This study was supported in part by Ellison Medical Foundation Senior Scholar Award for Aging (AG-SS-2482-10 to M.J.W.) and by NIH research grants (S10OD025132, S10OD028504, R01CA239165, R01AG072562 to M.-M.Z.).
Conflict of Interest
M.-M.Z. is a founder, director, and shareholder of Parkside Scientific, Inc.
References
- Cooke Bailey JN, Pericak-Vance MA, Haines JL (2014) Genome-wide association studies: getting to pathogenesis, the role of inflammation/complement in age-related macular degeneration. Cold Spring Harb Perspect Med 4(12): a017186.
- Schwartz SG, Hampton BM, Kovach JL, Brantley MA (2016) Genetics and age-related macular degeneration: a practical review for the clinician. Clin Ophthalmol 10: 1229-1235.
- Rabin DM, Rabin RL, Blenkinsop TA, Temple S, Stern JH (2013) Chronic oxidative stress upregulates Drusen related protein expression in adult human RPE stem cell-derived RPE cells: a novel culture model for dry AMD. Aging (Albany NY) 5(1): 51-66.
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al. (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308(5720): 385-389.
- Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, et al. (2005) Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet 77(1): 149-153.
- Sobrin L, Reynolds R, Yu Y, Fagerness J, Leveziel N, et al. (2011) ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am J Ophthalmol 151(2): 345-352 e3.
- Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, et al. (2010) Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci USA 107(16): 7395-7400.
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, et al. (2010) Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci USA 107(16): 7401-7406.
- Weber BH, Vogt G, Pruett RC, Stohr H, Felbor U (1994a) Mutations in the tissue inhibitor of metalloproteinases3 (TIMP3) in patients with Sorsby's fundus dystrophy. Nat Genet 8(4): 352-356.
- Weber BH, Vogt G, Wolz W, Ives EJ, Ewing CC (1994b) Sorsby's fundus dystrophy is genetically linked to chromosome 22q13-qter. Nat Genet 7(2): 158-161.
- Teper SJ, Nowinska A, Pilat J, Palucha A, Wylegala E (2010) Involvement of genetic factors in the response to a variable-dosing ranibizumab treatment regimen for age-related macular degeneration. Mol Vis 16: 2598-2604.
- Seddon JM, Reynolds R, Rosner B (2009) Peripheral retinal drusen and reticular pigment: association with CFHY402H and CFHrs1410996 genotypes in family and twin studies. Invest Ophthalmol Vis Sci 50(2): 586-591.
- Seddon JM, Silver RE, Kwong M, Rosner B (2015) Risk Prediction for Progression of Macular Degeneration: 10 Common and Rare Genetic Variants, Demographic, Environmental, and Macular Covariates. Invest Ophthalmol Vis Sci 56(4): 2192-2202.
- Awh CC, Hawken S, Zanke BW (2015) Treatment response to antioxidants and zinc based on CFH and ARMS2 genetic risk allele number in the Age-Related Eye Disease Study. Ophthalmology 122(1): 162-169.
- Juel HB, Faber C, Udsen MS, Folkersen L, Nissen MH (2012) Chemokine expression in retinal pigment epithelial ARPE-19 cells in response to coculture with activated T cells. Invest Ophthalmol Vis Sci 53(13): 8472-8480.
- Meyers KJ, Liu Z, Millen AE, Iyengar SK, Blodi BA, et al. (2015) Joint Associations of Diet, Lifestyle, and Genes with Age-Related Macular Degeneration. Ophthalmology 122(11): 2286-2294.
- Wang JJ, Buitendijk GH, Rochtchina E, Lee KE, Klein BE, et al. (2014) Genetic susceptibility, dietary antioxidants, and long-term incidence of age-related macular degeneration in two populations. Ophthalmology 121(13): 667-675.
- Vavvas DG, Small KW, Awh CC, Zanke BW, Tibshirani RJ (2018) CFH and ARMS2 genetic risk determines progression to neovascularization in age related macular degeneration after antioxidant and zinc supplementation. Clinical Trial 115(4): E696-E704.
- Giansanti V, Rodriguez GE, Savoldelli M, Gioia R, Forlino A, et al. (2013) Characterization of stress response in human retinal epithelial cells. J Cell Mol Med 17(1): 103-115.
- Parmeggiani F, Sorrentino FS, Romano MR, Costagliola C, Semeraro F, et al. (2013) Mechanism of inflammation in age-related macular degeneration: an up to date on genetic landmarks. Mediators Inflamm 2013: 435607.
- Ugurlu N, Asik MD, Yulek F, Neselioglu S, Cagil N (2013) Oxidative stress and anti-oxidative defence in patients with age-related macular degeneration. Curr Eye Res 38(4): 497-502.
- Handa JT, Reiser KM, Matsunaga H, Hjelmeland LM (1998) The advanced glycation endproduct pentosidine induces the expression of PDGF-B in human retinal pigment epithelial cells. Exp Eye Res 66(4): 411- 419.
- Handa JT, Verzijl N, Matsunaga H, Aotaki-Keen A, Lutty GA, et al. (1999) Increase in the advanced glycation end product pentosidine in Bruch's membrane with age. Invest Ophthalmol Vis Sci 40(3): 775-779.
- Mitter SK, Song C, Qi X, Mao H, Rao H, et al. (2014) Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy 10(11): 1989-2005.
- Cano M, Wang L, Wan J, Barnett BP, Ebrahimi K, et al. (2014) Oxidative stress induces mitochondrial dysfunction and a protective unfolded protein response in RPE cells. Free Radic Biol Med 69: 1- 14.
- Perkins ND (2007) Integrating cell-signaling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8: 49-62.
- Sporn MB, Liby KT (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564-571.
- Lambros ML, Plafker SM (2016) Oxidative Stress and the Nrf2 Antioxidant Transcription Factor in AgeRelated Macular Degeneration. Adv Exp Med Biol 854: 67-72.
- Sachdeva MM, Cano M, Handa JT (2014) Nrf2 signaling is impaired in the aging RPE given an oxidative insult. Exp Eye Res 119: 111-114.
- Eichenbaum JW, Cinaroglu A, Eichenbaum KD, Sadler KC (2009) A zebrafish retinal graded photochemical stress model. J Pharmocol Toxicol Methods 59: 121-127.
- Sanchez R, Zhou MM (2009) The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel 12: 659-665.
- Ghosh S, Chin CKK (2016) Analysis of RNA Seq Data using Top Hat and Cufflinks. Methods Molec Biol 1374: 339-361.
- Lawson ND, Li R, Shin M, Grosse A, Onur Yukselen, et. al. (2020) An improved zebrafish transcriptome annotation for sensitive and comprehensive detection of cell type specific genes. Elife 9: e55792.
- Nudelman G, Frasca A, Kent B, Sadler KC, Sealfon SC, et. al. (2018) High resolution of annotation of zebrafish transcriptome using long read sequencing. Genome Research 28: 1415-1425.
- Li H, Durbin R (2010) Fast and Accurate long-read alignment with Burrows Wheeler transform. Bioinformatics 26(5): 589-595.
- Wang L, Feng Z, Wang Xi, Wang Xiaowo, Zhang X (2010) DeGseq: and R package for identifying differential expressed genes from RNAseq data. Bioinformatics 26(1): 136-138.
- Mukhopadhyay P, Rajesh M, Yoshihiro K, Hasko G, Pacher P (2007) Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem Biophys Res Commun 358: 203-208.
- Ambivero CT, Cilenti L, Main S, Zervos AS (2014) Mulan E3 ubiquitin ligase interacts with multiple E2 conjugating enzymes and participates in mitophagy by recruiting GABARAP. Cell Signal 26: 2921-2939.
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299-334.
- Sarikas A, Hartmann T, Pan ZQ (2011) The cullin protein family. Genome Biol 12(4): 220.
- Villeneuve NF, Lau A, Zhang DD (2010) Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal 13: 1699-1712.
- Perkins ND (2007) Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8: 49-62.
- Millen AE, Meyers KJ, Liu Z, Engelman CD, Wallace RB, et al. (2015) Association between vitamin D status and age-related macular degeneration by genetic risk. JAMA Ophthalmol 133: 1171-1179.
-
Chih-Hung Chen, SiDe Li, Elena Rusinova, Guangtao Zhang, Joseph W Eichenbaum, Martin J. Walsh and Ming-Ming Zhou.Inhibition of BRD4 Attenuates Inflammation and Oxidative Stress in Retina. W J Opthalmol & Vision Res. 4(2): 2022. WJOVR.MS.ID.000584. DOI: 10.33552/WJOVR.2022.04.000584.
-
Bromodomain inhibitor, VEGF inhibitor, Retinal degeneration, Transcription, Oxidative stress, Autophagy, Nrf2, Mul1b, Mitochondria, Apoptotsis, Excalibur Pathology Inc, Norman, Oklahoma, Utraviolet light
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






