 Research Article
Research Article
Could autosomal dominant TGFBI-related corneal dystrophies be modelled in zebrafish by using CRISPR/Cas9: Challenges and Possibilities
Yaylacıoğlu Tuncay Fulya1,2*, and Dinçer Pervin Rukiye1
1Hacettepe University Faculty of Medicine, Department of Medical Biology, Ankara, Turkey
2Health Institutes of Turkey, Biotechnology Institute, Ankara, Turkey
Fulya Yaylacıoğlu Tuncay, Hacettepe University Faculty of Medicine, Department of Medical Biology, Ankara, Turkey.
Received Date: December 19, 2018; Published Date: January 16, 2019
Abstract
TGFBI-related corneal dystrophies are disorders characterized by accumulation of the abnormal protein product of TGFBI gene in the nonamyloid or amyloid form due to missense gain of function mutations of this gene. TGFBI gene exists in the zebrafish genome without duplication and has 65% homology to its human ortholoque. Arginine residue on the 124th position of the protein product of this gene was conserved in zebrafish. According to these data; in this study we tried to knock-in one of the gains of function mutations in the region coding 124th aminoacid residue of TGFBI gene in the zebrafish genome by using CRISPR/Cas9- mediated homology dependent repair method that resulted in many challenges in zebrafish. Finally, we could not achieve knockin of the precise mutation in the target sequence despite using several possibilities regarding CRISPR/Ca9 technique, but we could achieve in/del variations at the target sequence that could mimick the pathogenesis if the variation results in inframe change affecting the target arginine residue.
Keywords: CRISPR/Cas9; TGFBI; Knockin; Zebrafish; Corneal dystrophy; Genome editing
Introduction
TGFBI-related corneal stromal dystrophies are a group of hereditary corneal diseases caused by the mutations of the TGFBI gene [1]. Reis–Bücklers corneal dystrophy (RBCD), Thiel– Behnke corneal dystrophy (TBCD) are included in this group and mainly affect the Bowman layer of the cornea. Granular corneal dystrophy type I (GCD1), granular corneal dystrophy type II (GCD2), lattice corneal dystrophy type I (LCD1) and variant lattice dystrophies are also included in this group, but they mainly affect the stromal layer of the cornea. TGFBI gene is located on chromosome 5q31 and has 17 exones [2]. The protein product of this gene is keratoepithelin (TGFBIp) which is an extracellular matrix protein that is found in many tissues, but mutations in this gene cause production of abnormal protein causing progressive deposits detected only in the cornea [3]. Although several mutations from different countries were reported in this gene, there are two mutational hot spots, corresponding to arginine residues at positions 124 and 555 located in exons 4 and 12 of TGFBI, respectively [4,5]. Five genotype–phenotype correlations were mainly reported in these two hot spots: R124C in LCDI, R124H in ACD, R124L in RBCD, R555Q in TBCD and R555W in GCD1 [4]. The exact underlying mechanisms causing these different phenotypes has not been revealed yet. Additionally, there are no animal models showing the similar phenotypical properties as seen in patients [6].
Zebrafish may be used as an animal model for this group of disorders because the structure and the content of the human and zebrafish cornea are very similar [7,8]. TGFBI gene exist in the zebrafish genome without duplication and has 65% homology to its human ortholoque, arginine residue on the 124th position of the protein product of this gene was conserved in zebrafish and the corneal expression of TGFBI was shown from the 24th hour postfertilization in zebrafish [9]. Additionally, the genome editing methods, especially CRISPR/Cas9 system can be efficiently and easily used in zebrafish to mimick genetic diseases.
The latest discovered genome-editing platform is the CRISPR (Clustered regularly interspaced short palindromic repeat) and it is associated with Cas9 protein. The two components of this system (crRNA and tracrRNA) were fused into a single component namely single guide RNA (sgRNA) and this system was shown to recognize different sequences by only changing the sgRNA [10]. This system has been widely introduced to eukaryotic organisms such as yeasts plants and mammals to create targeted DSBs successfully. DNA double strand break (DSB) occurs by the recognition of the target sequence by sgRNA and the binding of the Cas9 protein to the protospacer adjacent motif (PAM) sequence downstream of the gRNA target sequence [11]. There exist different types of Cas9 proteins obtained from different microrganisms [12,13] or generated by engineering existing Cas9 proteins with different PAM specificities to increase the targetting capacity of CRISPR systems [14]. After the formation of the DSB by the CRISPR/ Cas9, there are two major DNA repair mechanisms to protect the integrity of the genome: 1) Homology-directed repair (HDR) and 2) Nonhomologous end-joining (NHEJ) [15]. NHEJ occurs by rejoining the two broken ends of the genome in a process that often results in small insertions and/or deletions at the site of the cleavage. If there is a donor template during the repair process, HDR can occur. Normally, when living cells are exposed to genotoxic injury, homologous sister chromatids are used as a template for HDR in replicating cells. In the case of genome editing of an extrachromosomal donor sequence, plasmid or single strand oligonucleotide can be used to integrate sequences of choice which are adjacent to induced DSBs. In cells, NHEJ pathway is much more efficient than HDR which makes it difficult to achieve precise changes in the genome [16]. The researchers mainly face the problems of efficiency in precise sequence modification methods such as gene insertion or gene correction where HDR is the main rate-limiting step.
According to these data our aim was to generate variation in the region coding 124th aminoacid residue of TGFBI gene with CRISPR/ Cas9 technology. These variations could be generated in two ways by using this tools: If this variation is generated by homologydirected repair mechanism, it could be possible to change the 124th aminoacid residue of arginine with systeine, leucine, histidine or serine. In literature the success rate of this mechanism was reported as 0-8% [17]. If the targeted mutation could not be generated with this mechanism, the more frequent mechanism of non-homology end-joining will occur, and this will create small insertions and deletions in the target sequence. From these mutations the ones that would not result in frameshift mutation, stop codon formation and also generate variations around the targeted site may cause accumulation of the protein in the corneal stroma like human corneal dystrophies.
Materials and Methods
SgRNA target site design
For the target region in the tgfbi gene the ENSDARG0000071586 coded reference sequence is used. Additionally, the target sequence is controlled in the wild type TU-AB zebrafish by Sanger sequencing in order to detect any variations in this region (Figure 1). CRISPRscan (http://www.crisprscan.org), CRISPRdirect (http://crispr.dbcls. jp/) tools were used to design sgRNAs for SpCas9 and CasBLASTR (http://www.CasBLASTR.org) tool was used to design sgRNA for Cas9 VQR variant. Alternative sgRNA formulations that were shown to be efficient previously [18] and could be synthesized by either SP6 or T7 RNA polymerase were designed in order to generate the DSB closer to the target region. The designed oligonucleotide sequences for the synthesis of the sgRNA mRNAs were listed in (Table 1).

SgRNA synthesis
For sgRNA synthesis, cloning-free, oligo-based method was used [19]. For this method in addition to target-specific DNA oligo, a universal oligo for the guide RNA was used (Table 1). The target-specific oligo contains the either T7 or SP6 promoter and the 15-20 nucleotide target sequence and a 20nucleotide sequence complementary to the guide RNA. The universal oligo and the target-specific oligos are annealed and extended with DNA polymerase and the resulting products after PCR purification by using PCR purification kit (Qiagen) were used as a template for in vitro transcription (IVT). For IVT reaction AmpliScribe T7-Flash transcription kit (Epicenter) was used for T7 and ‘MaxiScript SP6 Transcription (Invitrogen) kit was used for SP6 as described by Burgess et al. [19].
Table 1: Primers used to generate DNA IVT templates (sgRNA).
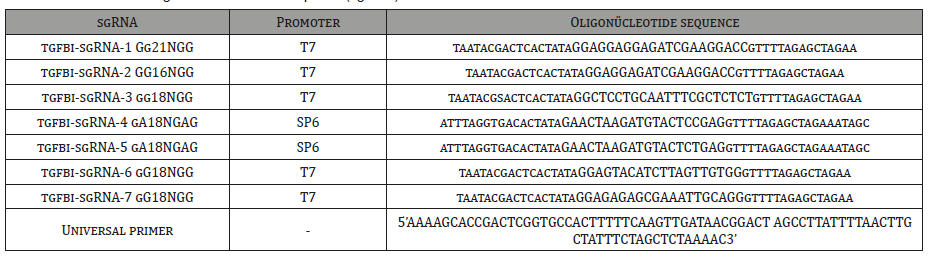
Transcription (Invitrogen) kit was used for SP6 as described by Burgess et al. [19]. After IVT, sgRNAs were treated with DNase and precipitated with Sodium Acetate/Ethanol. SgRNAs were checked in a %2 agarose gel for RNA integrity, then concentrations and A260/280 ratios were measured. For multiplex sgRNA synthesis, the equimolar concentrations of each target specific oligo (10μM of each) were mixed before annealing reaction as previously described.
Cas9 and VQR variant Cas9 mRNA synthesis
PT3TS-nls-zCas9-nl plasmid was used for the synthesis of Cas9 ve Cas9 VQR variant. After the linearization of the plasmid DNA with XbaI, mRNA was synthesized using mMessage mMachine T3 Transcription Kit (Invitrogen). Then, mRNAs were treated with DNAse and purified by using the RNeasy mini kit (Qiagen). Recombinant Cas9 protein (PioBNA) was also used instead of Cas9 mRNA in the microinjections [20].
SsODN design
In order to achieve precise nucleotide changes on the target sequence, single-stranded oligodeoxynocleotides (ssODNs) were designed (Table 2). Two ssODNs were designed (tgfbissODN-t and tgfbi-ssODN-nt) complementary to either the nontarget strand or the target strand. The homology arms of the ssODNs were designed asymmetrically in relation to the cut site: longer homology arm distal to the PAM site. [17]. Both ssODNs contain silent nucleotide changes near the PAM site to prevent sgRNA to bind and cut the repaired DNA. Additionally, the designed ssODNs contain the targeted missense variation to achieve the desired change in the amino acid sequence of the TGFBIp and the ApaI restriction ezyme cut site (5’ GGGCCC 3’) to permit screening by restriction fragment length polymorhism method (RFLP) after the microinjections. The designed ssODNs were ordered as ultra-purified products (SIGMAALDRICH).
Table 2: SsODN sequences.

Zebrafish embryo injections
Zebrafish embryos were obtained from breeding of wild type TU-AB zebrafish, all the animals were maintained at 25°C under a 12/12-hour light/ dark cycle in Hacettepe Zebrafish Research Laboratory located in Ankara, Turkey. The research protocol was approved by Hacettepe Animal Experimentations Local Ethics Board. The injection solution was prepared that contains the Cas9 mRNA (200-400 ng/μl) and gRNA (30-60 ng/μl) or Cas9 (200- 400 ng/μl) and gRNA (30-60ng/μl) and ssODN (60 ng/μl) or Cas9 protein (200 ng/μl) and gRNA (30-60ng/μl) and ssODN (60 ng/ μl). The injections were done to one-cell stage zebrafish eggs as a total volume of 1.5 nl into each embryo. Both the injected and control embryos were kept in 29°C and evaluated at 24th hour post fertilization (hpf). Dead deformed, and live embryos were recorded (Table 3).
Screening of the injected embryos
For each microinjection group, injected zebrafish embryos at 36 hpf (n=12-32) were collected and genomic DNA was extracted using the following technique: Embryos were incubated in a 50μl of 100mM of NaOH at 95 °C for 20 minutes. Then, 20 μl of Tris-HCl 1 M ph 7.5 was added (12). 3μl of this DNA were used in PCR with the following primers:
Tgfbi-forward: CAGTAAGAGCACTGGGTCAAGA
Tgfbi-reverse: AGACGCATTGGGAACTCACAG
Amplified PCR product of 298 base pairs for the target region were evaluated by T7 Endonuclease Enzyme (T7EI) assay for the evaluation of the presence of mosaicism [21]. For the HDR evaluation, RFLP assay was used with ApaI restriction enzyme. And all the samples were re-evaluated by Sanger sequencing for the presence of mosaicism in the target sequence.
Result
Finding the sgRNA that has the higher DSB formation ability closer to the target
The main aim of this study is to create a precise missense variation in the defined sequence. To achieve this goal NHEJ which is the predominant repair mechanism in cells when there is a DSB, is not useful. HDR is the only repair mechanism that is able to create the target missense variation. However, we first should have a sgRNA/Cas9 pair that cuts the defined sequence efficiently and closer to the target sequence. As soon as we defined this, we could try microinjections with ssODN to achieve HDR in zebrafish embryos.
The screening results of embryos after microinjections with designed sgRNAs and Cas9 mRNA or sgRNAs and Cas9 VQR variant mRNA was shown in Table 3 & Figure 2. The highest DSB formation ability in T7EI assay and Sanger sequencing was shown in the injection group 2 in which tgfbi-sgRNA-2 and Cas9 mRNA were used (Figure 2a1, 2a2). When we used this sgRNA the DSB occured 26 bp distal to the target sequence which could decrease the efficiency of HDR in the following experiments.
In the third injection group, target sequence of the tgfbisgRNA- 3 was followed by the PAM site that includes the polymorphic region in the wild-type zebrafish that was shown in (Figure 1). Additionally, it has two mismatches at the 5’ region (gg18NGG) that decreased the expected efficiency. As a result, the screening with T7EI assay showed no positive results in this injection group (Table 3; Figure 2b1, 2b2)
Table 3: Injection groups and screening results.
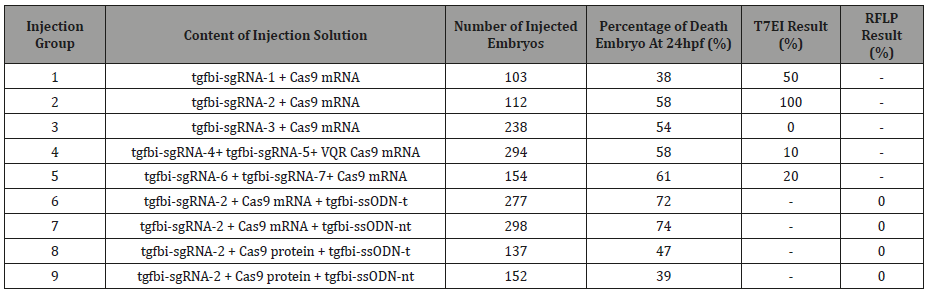
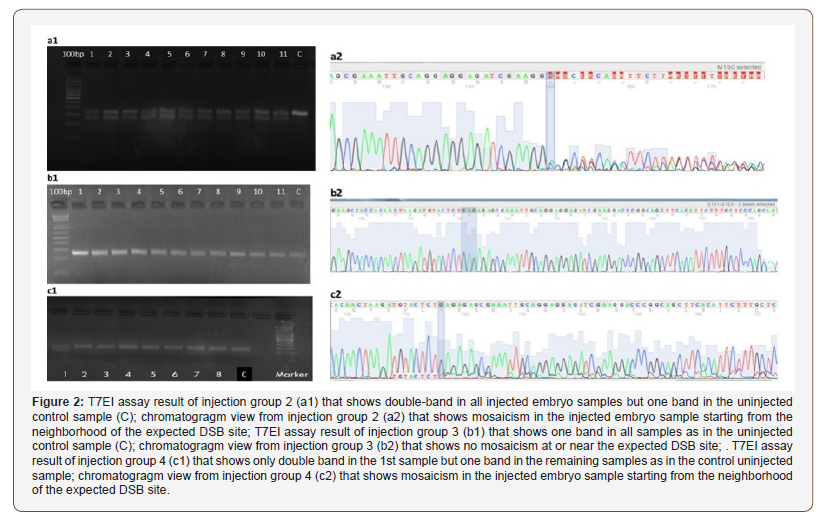
In the injection groups 4 and 5 the DSB locations were expected to be closer to the target sequence but the DSB formation efficiency in these groups was found to be very low (1020%) (Table 3; Figure 2c1,2c2).
Evaluation of the Cas9-mediated HDR efficiency
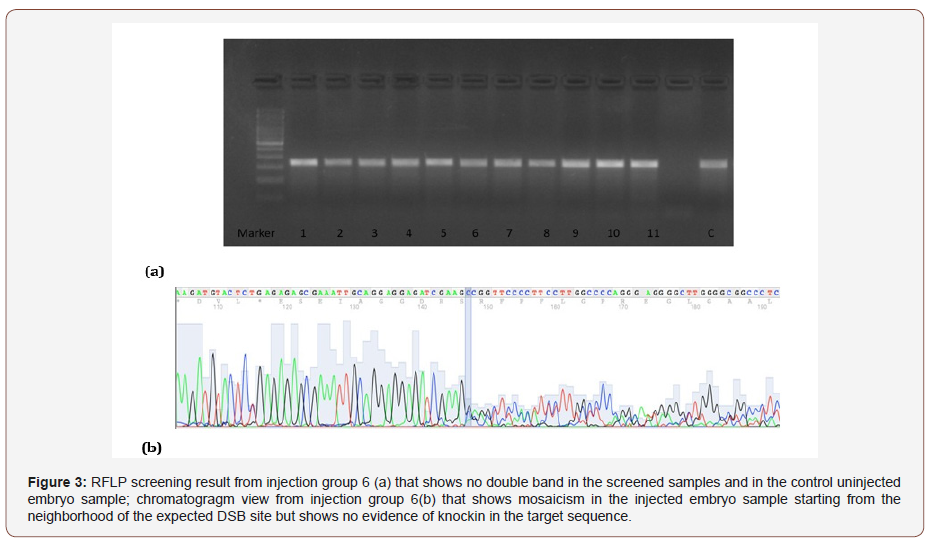
According to the previous results the microinjections were planned using tgfbi-sgRNA-2 and Cas9 mRNA or Cas9 protein with ssODNs to achieve HDR. The screening results using RFLP and Sanger sequencing were shown in (Table 3 & Figure 3).
The mortality rate was found to be higher in the injection groups where Cas9 mRNA was used. However, the mortality rates were found to be less in the groups where Cas9 protein was used (Table 3). The HDR that will create a missense variation in the target region and a AsaI RE site formation could not be shown in the screened embryos by RFLP and Sanger sequencing (Figure 3).
Discussion
In this study, first we tried several CRISPR/Cas9 options in order to achieve DSB at or close to our target sequence and secondly, we tried to achieve Cas9-mediated HDR to obtain desired missense change at the target sequence to mimick the TGFBIrelated corneal dystrophies in zebrafish. Although there have been several studies and reports in the CRISPR/Cas9 genome-editing technique for decreasing the limitations of targetting, achieving precise changes especially in zebrafish genome by HDR is still low and highly variable.
During the sgRNA design, we had two limitations in both ends of the target sequence: First limitation is the existence of 5’GG for T7 promoter or 5’GA for SP6 promoter in the target and secondly existence of PAM sequence (3’NGG) for Cas9. In the exon 4 of tgfbi of zebrafish, there exists no sequence compatible with these two limitations. However, the study of Giraldez et al. reported that they could extend the CRISPR target repertoire with truncated, extended and 5ʹ mismatch-containing sgRNAs that allowed us to design several sgRNAs around our target sequence [18]. In accordance with this report, the most efficient sgRNA was shown to be the tgfbi-sgRNA-2 as having no mismatch but truncated to 16 nucleotide instead of 18 (GG16). The cutting efficiency of this sgRNA was found to be 100% in the screening of injected embryos whereas the cutting efficiency of the sgRNA with one mismatch and 20 nucleotide was shown to be 40% (Table 3).
For achieving Cas9-mediated HDR, there are three important factors affecting the efficiency: The efficiency of DSB formation of the CRISPR/Cas9, the distance of the DSB to the target sequence and the properties of the donor template [17,22]. The most important disadvantage of our most efficient sgRNA was the distance of the DSB to the target site which was 26 nucleotide. It was mentioned in a report that the HDR efficiency is higher when the distance of DSB is not further than 10-20 nucleotide [17]. As a result we could not show any HDR event in the screening of any injection groups in which tgfbi-sgRNA-2 was used. In order to overcome this distance issue, we designed other sgRNAs closer to the target site that could be synthesized by SP6 promoter and used with VQR variant Cas9. Additionally, to overcome polymorphism problem seen in wild type zebrafish near the target sequence we synthesized sgRNA as multiplex including both sequence variations. But the efficiency in this group of injections remained to be 10%, too low for Cas9- mediated HDR. As a third option, we tried multiplex genome editing: designed sgRNAs targetting downstream and upstream of the target sequence; injected them together with ssODN. However, we could not show any HDR event. This could be resulted from that the simultaneous DSB formation efficiency of this system was found to be low (10 %).
The 24 hpf mortality rates were found to be high in the injection groups 6 and 7 in which injection solution included sgRNA, Cas9 mRNA and ssODN (72-74%) so higher concentrations could not be used in order to increase HDR efficiency. To overcome this problem, we used Cas9 protein instead of Cas9 mRNA in injection groups 8 and 9. In literature it was shown that Cas9 protein increased the HDR efficiency itself and decreased toxicity to injected embryos [20]. In our injection groups the mortality rate could be decreased relevant to the Cas9 mRNA (3947%), but the screening could not reveal any HDR event similar to the group 6 and 7.
In this study we could not achieve the Cas9-mediated HDR in the target sequence of tgfbi gene to mimick the R124 mutation in zebrafish. We could achieve in/del mutations at the target site in injection group 4 with VQR variant Cas9 and tgfbi-sgRNA-4 and 5 which could produce inframe changes affecting Arginine residue in the protein product and could give us a chance to mimick corneal changes as in human disorders. In addition to this option, if we could increase the DSB formation efficiency of injection groups 4 and 5 more than 50% we could try coinjection with a newly designed ssODN for these sgRNAs. Another option we could do in future trials is the usage of LpCpf1 protein which was shown to have higher HDR efficiency than SpCas9 protein [12]. Consequently, in zebrafish mimicking autosomal dominant disease by using CRISPR/ Cas9 mediated HDR has not been as easy as mimicking autosomal recessive diseases by using CRISPR/Cas9 mediated NHEJ despite the several improvements in this genome editing platform.
Acknowledgment
This study was supported by Hacettepe University Scientific Research Unit (Grant no: TAY-2017-12735).
Conflicts of Interest
No conflicts of interest.
References
- Weiss JS, Møller HU, Lisch W, Aldave AJ, Seitz B, el al. (2015) IC3D classification of the corneal dystrophies- Edition 2. Cornea 34(2): 117- 159.
- Escribano J, Hernando N, Ghosh S, Crabb J, Coca-Prados M (1994) cDNA from human ocular ciliary epithelium homologous to beta ig-h3 is preferentially expressed as an extracellular protein in the corneal epithelium. J Cell Physiol 160(3): 511-521.
- El Kochairi I, Letovanec I, Uffer S, Munier FL, Chaubert P, el al. (2006) Systemic investigation of keratoepithelin deposits in TGFBI/BIGH3- related corneal dystrophy. Mol Vis 12: 461-466.
- Munier FL, Frueh BE, Othenin Girard P, Uffer S, Cousin P (2002) BIGH3 mutation spectrum in corneal dystrophies. Invest Ophthalmol Vis Sci 43(4): 949-954.
- Han KE, Choi SI, Kim TI, Maeng YS, Stulting RD, et al. (2016) Pathogenesis and treatments of TGFBI corneal dystrophies. Prog Retin Eye 50: 67-88.
- Yamazoe K, Yoshida S, Yasuda M, Hatou S, Inagaki E, et al. (2015) Development of a Transgenic Mouse with R124H Human TGFBI Mutation Associated with Granular Corneal Dystrophy Type 2. PLoS One 10(7): e0133397.
- Akhtar SI, Schonthaler HB, Bron AJ, Dahm R (2008) Formation of stromal collagen fibrils and proteoglycans in the developing zebrafish cornea. Acta Ophthalmol 86(6): 655-665.
- Soules KA, Link BA (2005) Morphogenesis of the anterior segment in the zebrafish eye. BMC Dev Biol 5: 12.
- Zhao XC, Yee RW, Norcom E, Burgess H, Avanesov AS, et al. (2006) The zebrafish cornea: structure and development. Invest Ophthalmol Vis Sci 47(10): 4341-4348.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816-821.
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, et al. (2011) Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18(5): 529-536.
- Moreno Mateos MA, Fernandez JP, Rouet R, Lane MA, Vejnar CE, et al. (2017) CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun 8(1): 2024.
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520(7546): 186-191.
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, et al. (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523(7561): 481-485.
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, et al. (1998) Homologous recombination and non-homologous end joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 17(18): 5497-5508.
- Mao Z, Bozzella M, Seluanov A, Gorbunova V (2008) Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7(10): 1765-1771.
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34(3): 339- 344.
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, et al. (2015) CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12(10): 982-988.
- Varshney GK, Carrington B, Pei W, Bishop K, Chen Z, et al. (2016) A high-throughput functional genomics workflow based on CRISPR/Cas9- mediated targeted mutagenesis in zebrafish. Nat Protoc 11(12): 2357- 2375.
- Kotani H, Taimatsu K, Ohga R, Ota S, Kawahara A (2015) Efficient Multiple Genome Modifications Induced by the crRNAs, tracrRNA and Cas9 Protein Complex in Zebrafish. PLoS One 10(5): e0128319.
- Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, et al. (2015) Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS One 10(8): e0136690.
- Armstrong GA, Liao M, You Z, Lissouba A, Chen BE, et al. (2016) Homology Directed Knockin of Point Mutations in the Zebrafish tardbp and fus Genes in ALS Using the CRISPR/Cas9 System. PLoS One. 11(3): e0150188.
-
Yaylacıoğlu Tuncay Fulya, Dinçer Pervin Rukiye. Could autosomal dominant TGFBI-related corneal dystrophies be modelled in zebrafish by using CRISPR/Cas9: Challenges and Possibilities. W J Opthalmol & Vision Res. 1(3): 2019. WJOVR.MS.ID.000513.
-
CRISPR/Cas9, TGFBI, Knockin, Zebrafish, Corneal dystrophy, Genome editing, Hereditary corneal, Granular corneal dystrophy, Lattice corneal dystrophy, Keratoepithelin, Homology-directed repair.
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






