 Review Article
Review Article
Review on Methanogenesis and its Role
Mebrate Getabalew1, Tewodros Alemneh*2 and Etagegnehu Bzuneh3
1,3College of Agricultural and Natural Resources Science, Department of Animal Science, Debre Berhan University, Ethiopia
2Woreta City Office of Agriculture and Environmental Protection, South Gondar Zone, Amhara Regional State, Ethiopia
Tewodros Alemneh, Expert Veterinarian at Woreta City Office of Agriculture & Environmental Protection, South Gondar Zone, Amhara Regional State, Ethiopia.
Received Date: July 15, 2020; Published Date: December 07, 2020
Abstract
Methane (CH4) is the second major gas, next to CO2, responsible for the warming of environment and ozone layer depletion. It is a potent greenhouse gas with a global warming potential 25 times more than carbon dioxide. Methanogenesis is the biological production of methane mediated by anaerobic microorganisms from the domain Archaea commonly called methanogens. These methanogens are different from bacteria and eukarya as they lack peptidoglycan in their cell wall, which is present in bacteria and eukarya. Methane is produced by three major pathways on the basis of substrate utilized for methane production: hydrogenotropic, acetoclastic and methylotropic. Out of these, hydrogenotropic and acetoclastic are the two predominant pathways. There are two major sources of methane; namely, natural and anthropogenic. The natural sources include wetlands, termites and oceans, whereas, the common anthropogenic sources are fossil fuel transport and distribution, livestock, rice fields, and landfills. Among anthropogenic sources of methanogenesis, livestock which is ruminal methanogenesis is the major and it contribute to global warming and gross energy feed intake loss about 10-12%. The role of methanogenesis to ruminant animals is to remove hydrogen (H2) from the rumen. Methanogenesis can also beneficially exploit to treat organic wastes to produce useful compounds and methane that can be collected as biogas.
Keywords: Acetoclastic; Anthropogenic; Hydrogenotropic; Methane; Methanogenesis; Methylotropic
Introduction
Methanogenesis is a multi-step process involving different group of microorganisms like hydrolytic, fermentative, acetogenic and above all methanogenic micro-organisms. It is the biological production of methane mediated by anaerobic microorganisms from the domain Archaea commonly called methanogens. These methanogens are organism carrying out methanogenesis, requiring completely anaerobic conditions for growth. These methanogens are different from bacteria and eukarya as they lack peptidoglycan in their cell wall, which is present in bacteria and eukarya [1]. Methane is produced by three major pathways on the basis of substrate utilized for methane production: (1) hydrogenotropic (2) acetoclastic and (3) methylotropic. Out of these, hydrogenotropic and acetoclastic is the predominant pathway. Hydrogenotrophic methanogens, which reduce CO2 to CH4, are responsible for the major part of rumen methanogenesis [2].
Methane is one of the three main greenhouse gases, together with CO2 and nitrous oxide (N2O), its global warming potential is 25- fold than that of CO2. CH4 also affects the degradation of the ozone layer [3]. Men are responsible for about two third of the total global CH4 emission called total anthropogenic methane [4]. Agriculture accounts for 47-56% of total anthropogenic CH4 emissions [5,6]; of this amount may be 12-37% of enteric origin [7].
Human-related methane emissions are mainly produced by domestic ruminants, rice field, carbon mines, landfills, and fossil fuel usage [4]. On the other hand, methane also emitted from natural source such as from wetland, oceans and termites [1]. Among animals, ruminants are the primary emitters of CH4. Their rumen, a large fore stomach, has a continuous fermentation system. The rumen occupies more than 70% of the total stomach capacity and its volume is 15 liters in sheep and 100-150 liters in cattle [5]. Methane production arises principally from microbial fermentation of hydrolyzed carbohydrates and is considered an energy loss for the animal [8]. Many factors influence ruminant CH4 production, including level of intake, type and quality of feeds, energy consumption, animal size, growth rate, level of production, genetics, and environmental temperature [5].
Methane emission from ruminants reduces the efficiency of nutrient utilization. Therefore, manipulation of rumen microbial ecosystem for reducing methane emission by ruminants to improve their performance is one of the most important goals for animal nutritionists. Reduction in methane emission from ruminants enhances the efficiency of nutrient utilization and augments productivity and also reduces methane impact on global warming [9]. There are some research reports about methanogen, and methanogenesis from ruminants and sources of methanogenesis; however, there is no organized information about methanogenesis and its role. Therefore, the objective of this review is to organize information on methanogenesis and its role as well as source of methanogenesis.
Methanogenesis: General Concepts
Methanogenesis is the formation of methane performed by methanogenic Archaea, a specialized group of microbes present in several anaerobic environments including oceans, marshes and swamps, rice paddies, geothermal habitats, anaerobic sewage digestion systems and animal gastrointestinal tracts. Biological methanogenesis plays a major role in the carbon cycle on earth. Methane escaping from anaerobic habitats can serve as a carbon and energy source for aerobic methanotrophic bacteria or can escape to the atmosphere, where it is a major participant in atmospheric chemical reactions and is an important greenhouse gas [10].
Methanogenesis or biomethanation is the formation of methane by microbes known as methanogens. Organisms capable of producing methane have been identified only from the domain Archaea, a group phylogenetically distinct from both eukaryotes and bacteria, although many live-in close association with anaerobic bacteria. The production of methane is an important and widespread form of microbial metabolism. In anoxic environments, it is the final step in the decomposition of biomass. Methanogenesis is responsible for significant amounts of natural gas accumulations, the remainder being thermogenic [11-13].
Biochemistry
Methanogenesis in microbes is a form of anaerobic respiration [14]. Methanogens do not use oxygen to respire; in fact, oxygen inhibits the growth of methanogens. The terminal electron acceptor in methanogenesis is not oxygen, but carbon. The carbon can occur in a small number of organic compounds, all with low molecular weights. The two best described pathways involve the use of acetic acid or inorganic carbon dioxide as terminal electron acceptors:
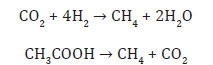
During anaerobic respiration of carbohydrates, H2 and acetate are formed in a ratio of 2:1 or lower, so H2 contributes only 33% to methanogenesis, with acetate contributing the greater proportion. In some circumstances, for instance, in the rumen, where acetate is largely absorbed into the bloodstream of the host, the contribution of H2 to methanogenesis is greater [15,16].

However, depending on pH and temperature, methanogenesis has been shown to use carbon from other small organic compounds, such as formic acid (formate), methanol, methylamines, tetramethylammonium, dimethyl sulfide, and methanethiol. The catabolism of the methyl compounds is mediated by methyl transferases to give methyl coenzyme M [14,16] (Figure 1).
Mechanisms and coenzymes for methanogenesis
The biochemistry of methanogenesis involves the following coenzymes and cofactors: F420, coenzyme B, coenzyme M, methanofuran, and methanopterin. The mechanism for the conversion of CH3-S bond into methane involves a ternary complex of methyl coenzyme M and coenzyme B fit into a channel terminated by the axial site on nickel of the cofactor F430. One proposed mechanism invokes electron transfer from Ni (I) (to give Ni (II)), which initiates formation of CH4. Coupling of the coenzyme M thiyl radical (RS.) with HS coenzyme B releases a proton and re-reduces Ni (II) by one-electron, regenerating Ni (I) [17].
In other words, methanogenesis pathways utilize several coenzymes of which methanofuran (MF), tetrahydromethanopterin (H4MPT), tetrahydrosarcinapterin (H4SPT) and coenzyme M (or HSCoM) carry the carbon moiety destined to generate methane, while coenzyme F420, coenzyme B (HS-CoB), methanophenazine and coenzyme F430 transfer electrons that are used in carbon reduction [1].
Methanogenesis pathway
The production of methane is through various pathways. The important pathways include the followings: (1) The predominant pathway is the hydrogenotrophic using CO2 as the carbon source and H2 as the main electron donor [18]. (2) Methane is also produced from acetate via the acetoclastic pathway e.g. Methanosacrcina and Methanotrix [1]. (3) Formate is also an important electron donor used by many rumen hydrogenotrophic methanogens and may account for up to 18% of the methane produced in the rumen [19]. (4) Methylamines and methanol produced in the rumen can also be used by methylotrophic methanogens of the order Methanosarcinales and Methanobacteriales [1]. All the pathways have in common the demethylation of methylcoenzyme M to methane and the reduction of the heterodisulfide of coenzyme-M and coenzyme-B catalyzed by methyl-coenzyme-M and heterodisulfide reductases [1].
Reverse methanogenesis
Some organisms can oxidize methane, functionally reversing the process of methanogenesis, also referred to as the anaerobic oxidation of methane (AOM). Organisms performing AOM have been found in multiple marine and freshwater environments including methane seeps, hydrothermal vents, coastal sediments and sulfatemethane transition zones [20]. These organisms may accomplish reverse methanogenesis using a nickel-containing protein similar to methyl-coenzyme-M reductase used by methanogenic Archaea [21]. Reverse methanogenesis occurs according to the reaction:
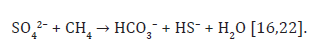
Sources of Methanogenesis
Methanogenesis in rice fields and wetlands
Wetlands and rice fields are characterized by water-logged soils and distinctive communities of plant and animal species that have evolved and adapted to the constant presence of water. Due to this high level of water saturation as well as warm weather, wetlands and rice fields are one of the most significant natural sources of atmospheric methane [1].
Anaerobic decomposition of organic material in flooded rice fields produces methane. Anaerobic conditions occur in rice field as a result of soil submergence. Water saturation of soil limits the transport of oxygen in soil resulting in higher activity of methanogens to produce methane. Under anaerobic and reduced conditions methanogens produce methane either by using CO2 and H2 or by using acetate. Under steady conditions methanogenesis by acetoclastic pathway predominates and accounts for 75-80% of total methane emitted [1].
Methanogenesis in termites
Termites have good sources of wood degrading enzymes such as xylanases, laccases; as their main dietary component is wood [23]. Along with bacterial species like Bacteroides Cellulomonas, Spiromusa termitida etc. termites harbour flagellate protists that fill up the bulk of the hindgut paunch. The gut flagellate includes Trichonympha, Calonympha which degrade the lignocellulosic feed with formation of excess hydrogen as intermediate. This large number of bacterial, archael and protozoal population inhabit the gut of termites. This partnership with a diverse community of bacterial, Archaeal and eukaryotic gut symbionts break down the plant fiber and ferment the products to acetate and variable amounts of methane, with hydrogen as a central intermediate [1].
The fermentation of wood polysaccharides by the gut flagellates yields acetate and other short-chain fatty acids, which are resorbed by the host. Hydrogen is an important intermediate that drives the reduction of CO2, which yields additional acetate and some methane. Although H2 may strongly accumulate at the gut center, most of it is consumed before it can escape from the gut. Methanogenic Archaea (methanogens) that inhabit the gut of termites generate enormous amount of methane that adds to the global atmospheric methane (CH4). The predominant species is Methanosarcina barkeri using acetate as major source of methane production. The total methane contribution due to termites is probably less than 15 Tg (Teragram) (1 Tg = 1 million tons) per year [24].
Methanogenesis in landfills
A mixture of organic and inorganic wastes is disposed at a landfill with varying humidity and much heterogeneity. Approximately 75% of municipal waste is biodegradable organic material. Substances in waste have various decomposition rates. Food waste is most readily degraded, paper, cardboard, wood and textile waste decompose slowly, while plastics and rubber are not degraded at all. A number of factors affect the quantity of gases formed at landfills and their composition, such as waste type and age, quantity and type of organic components, waste humidity and temperature [1].
Methanogenesis in oceans
A large amount of methane is generated in ocean sediments which travel a long way to get to the surface and escape to the atmosphere. Nitrosopumilus maritimus the most abundant microorganisms in marine surface waters contribute of oceans to total methane production about 10 Tg [25].
Methanogenesis from fossil fuels
Methane is naturally present in fossil fuels due to long term decomposition of organic matter. Methane from fossil-fuel production is primarily emitted through: (1) The combustion of extracted fossil-fuels; (2) The industry practices of venting, or intentionally releasing excess gas, and flaring, or intentionally burning excess gas; (3) Fugitive emissions, which include unintentional leakage from the transportation, storage, and distribution of fossil fuels. The combustion of fossil fuels mined, drilled, and otherwise extracted on federal lands and waters contributed approximately 62,000 metric tons (U.S.A) of methane to the atmosphere in 2012 alone, or more than 1.5 million metric tons of CO2 equivalents [1].
Methanogenesis in herbivores
Emissions from enteric fermentation of pigs and horses are of minor importance, but not negligible. Methane production is influenced in particular to diet composition and feeding practices. The formation of CH4 in the digestive system (enteric fermentation) of pig and horse is mainly centered in the hind gut (colon). Here, bacterial action degrades those organic species that passed the digestive tract undigested, mainly cellulose, hemi-cellulose and pectin which are summed up as bacterially fermentable substrates (BFS). Bacterial action converts these substrates to volatile fatty acids, CH4 and CO2. The fatty acids play an important role in the energy supply of pigs. In experiments with sows, about half the cellulose and about 90% of the sugar (xylose), starch 4 and cellulose (pectin) as well as the protein casein that were applied to the animals intra-caecally were degraded in the hind gut. The gross energy loss of feed in the form of methane is very less around 0.1-1% gross energy of feed intake. The emission of methane from horse and pig is 0.14 Tg and 1.7 Tg, respectively [26].
Methanogenesis in ruminants
Methane is produced in the rumen as a product of normal fermentation of feedstuffs. The major part of methanogenesis in ruminants occurs in the large fermentative chamber known as rumen [27]. In the rumen methanogens utilize hydrogen and carbon dioxide to produce methane but the Methanosarcina species are an exception because they grow slowly on these two substrates and therefore these species utilize methanol and methylamines to produce methane [28]. Although methane production can also occur in the lower gastro intestinal tract, as in non-ruminants 89% of methane emitted from ruminants is produced in the rumen and exhaled through the mouth and nose [1].
The methanogenesis process in the rumen is the last step in the anaerobic conversion of organic matter to methane. This entire course involves a large number of microorganisms. Bacterial species, fungi and protozoa hydrolyze the proteins, starch and plant cell wall polymers in amino acids and sugars. The amino acids and sugars are then fermented to volatile fatty acids (VFAs), hydrogen and carbon dioxide [29]. The VFA mainly acetate, propionate, and butyrate are used by the animal as source of energy while the gases are eliminated mainly through eructation. CO2 and H2 are using to form CH4, and thus reducing the metabolic H2 produced during microbial metabolism [30]. Fermentation is an oxidative process, during which reduced cofactors (NADH, NADPH, FADH) are re-oxidized (NAD-1, NADP-1, FAD-1) through dehydrogenation reactions releasing hydrogen in the rumen. As soon as produced, hydrogen is used by methanogenic Archaea, a microbial group distinct from Eubacteria, to reduce CO2 into CH4 [31]. Methanogens species use both hydrogen (80%) and formate (18%) to produce the methane gas [32].
Methanogenesis in humans
Some humans produce flatus that contains methane. In one study of the feces of nine adults, five of the samples contained Archaea capable of producing methane [30]. Similar results are found in samples of gas obtained from within the rectum. Even among humans whose flatus does contain methane, the amount is in the range of 10% or less of the total amount of gas [16].
Methanogenesis in plants
Many experiments have suggested that leaf tissues of living plants emit methane [33]. Other research has indicated that the plants are not actually generating methane; they are just absorbing methane from the soil and then emitting it through their leaf tissues [16].
Methanogenesis in soils
Methanogens are observed in anoxic soil environments, contributing to the degradation of organic matter. This organic matter may be placed by humans through landfill, buried as sediment on the bottom of lakes or oceans as sediments, and as residual organic matter from sediments that have formed into sedimentary rocks [34].
Methanogenesis in earth’s crust
Methanogens are a notable part of the microbial communities in continental and marine deep biosphere [16, 35-37].
Rumen Methanogens
Characteristics of methanogens
Methanogens are a sub-group of the Archaea and the phylum Euryarchaeota. Unlike Bacteria, methanogens lack peptidoglycan in the cell wall, replaced by pseudomurein in Methanobrevibacter and Methanobacterium, heteropolysaccharide in Methanosarcina, and protein in Methanomicrobium [1,38]. Methanogens possess unique cofactors such as coenzyme M, HSHTP, F420 and lipids important for methanogenesis process [29]. The F420 cofactor is necessary for the activity of hydrogenase and formate dehydrogenase enzymes and allows them to fluoresce blue-green at 420 nm [39]. Coenzyme-M acts as terminal methyl carrier in methanogenesis process and represent the smallest organic factor [40].
Among methanogens, the cell shape and characteristics also vary as well. The most important methanogen found in rumen, M. ruminantium is rod shaped with pseudomurein in the cell envelope and requires coenzyme M, hydrogen, carbon dioxide and formate for methane production [38]. From the same order (Methanobacteriales such as M. ruminantium) M. formicicum is non-motile rod or filament shaped with pseudomurein in the cell wall. The species that belong to Methanobacteriales and Methanomicrobiales orders are methanogens without cytochromes and their energy source is represented by hydrogen and formate [29]. The species from Methanosarcinales order are coccoid shaped without motility (and they have cytochromes). Cytochromes or membrane bound electron carriers, play a role in the oxidation of methyl group to carbon dioxide [41]. Methanosarcina spp. can use a large range of substrate such as H2, CO2, methanol, methylamines and acetate [38].
Methanoculleus olentangyi are present in the rumen in a large number in rumen liquor depending upon the type of diet given to animals, especially the fiber content in the ration. On a fiber rich diet, production of acetic acid is more coupled with more production of methane [42]. Rumen methanogens grow only in environments with a redox potential below 300 mV. More than sixty species were isolated from various anaerobic habitats like sanitary landfills, peat bogs, waterlogged soils, salt lakes, thermal environments, and intestinal tracts of animals. Only five of these species belonging to Methanobrevibacter and Methanosarcina genera, were isolated from rumen digesta. Only two of these species have been found at a population level greater than 106 ml-1L of rumen liquor [43].
Association of methagens with protozoa
Methanogens are known to have symbiotic relationships involving interspecies hydrogen transfer with rumen microorganisms, especially with rumen protozoa where the methanogens can be associated intracellularly and extracellularly. Common protozoa in the bovine rumen found to have such a relationship are from the genera Entodinium, Polyplastron, Epidinium, and Ophryoscolex, while the methanogens most often associated with protozoa are from the orders Methanobacteriales and Methanomicrobiales [44].
Factors affecting ruminal methanogenesis
There are ranges of factors that affect rumen methane emissions, such as feed intake, type of carbohydrate fermented, forage processing and lipid addition [45]. These factors have their effects by two different mechanisms. The first mechanism described is the amount of carbohydrate that is fermented in the reticulo-rumen. The second mechanism is the amount of available hydrogen and the consecutive methane formation through the ratio of Volatile Fatty Acids (VFAs) produced. The relation between the production of propionic and acetic acids has a relevant impact on methane production. The VFA’s regulate the hydrogen supply which controls the production of methane. If carbohydrate would be fermented to acetic acid only, the energy loss from methane formation would be 33% [46].
The Roles of Methanogenesis
Role of methanogenesis in ruminants
One role of methanogenesis in ruminant animals is that it helps in the removal of excessive hydrogen (H2) production in the rumen. A high hydrogen level in the rumen decreases carbohydrate degradation, decreases rate of microbial growth and thus decreases microbial protein synthesis essential for the body [1].
Role of methanogenesis in global warming
Methane is the second major gas after CO2 responsible for the warming of environment and ozone layer depletion. It is a potent greenhouse gas with a global warming potential 25 times more than carbon dioxide and thus methanogenesis in livestock and the decay of organic material is a considerable contributor to global warming) [16,47]. It may not be a net contributor in the sense that it works on organic material which used up atmospheric carbon dioxide when it was created, but its overall effect is to convert the carbon dioxide into methane which is a much more potent greenhouse gas. Methanogenesis can also be beneficiary exploited to treat organic waste, to produce useful compounds. It also be beneficially exploited to treat organic waste, to produce useful compound and the methane can be collected and used as biogas, a fuel [16,48]. It is the primary pathway whereby most organic matter disposed of via landfill is broken down [49].
Role of methanogenesis in extra-terrestrial life
The presence of atmospheric methane has a role in the scientific search for extra-terrestrial life. The justification is that methane in the atmosphere will eventually dissipate, unless something is replenishing it. If methane is detected (by using a spectrometer for example) this may indicate that life is, or recently was, present. This was debated [50] when methane was discovered in the Martian atmosphere by M.J. Mumma of NASA’s Goddard Flight Center, and verified by the Mars Express Orbiter (2004) [51] and in Titan’s atmosphere by the Huygens probe (2005) [52]. This debate was furthered with the discovery of ‘transient’, ‘spikes of methane’ on Mars by the Curiosity Rover [53].
It is also argued that atmospheric methane can come from volcanoes or other fissures in the planet’s crust and that without an isotopic signature, the origin or source may be difficult to identify [16,54,55].
On 13 April 2017, NASA confirmed that the dive of the Cassini orbiter spacecraft on 28 October 2015 discovered the Enceladus plume which has all the ingredients for methanogenesis-based life forms to feed from. Previous results, published in March 2015, suggested hot water is interacting with rock beneath the sea; the new findings support that conclusion and add that the rock appears to be reacting chemically. From these observations’ scientists have determined that nearly 98% of the gas in the plume is water, about 1% is hydrogen and the rest is a mixture of other molecules including carbon dioxide, methane and ammonia [16,56].
Conclusion
Methanogenesis is the formation of methane performed by methanogenic Archaea anaerobically. There are different sources of methanogenesis including both natural and anthropogenic (human sources). Among anthropogenic sources of methanogenesis, agriculture specially raising livestock is the main sources of methanogenesis. Methanogenesis in livestock production and decay of organic material is considerable contributor to global warming. It also is beneficially exploited to treat organic waste, to produce useful compound and methane that can be collected and used as biogas and a fuel. The role of methanogenesis in ruminant animal is to remove excessive hydrogen from the rumen as hydrogen accumulates excess it decrease carbohydrate degradation, decrease rate of microbial growth and thus decrease microbial protein synthesis.
Acknowledgement
None.
Conflict of Interest
No conflict of interest.
References
- Neeti L, Preeti L, Amir AS, Rakshanda B, Rouf RD, et al. (2017) Methanogenesis: Are ruminants only responsible: A review, Journal of Pharmacognosy and Phytochemistry 6(6): 2347-2352.
- Fonty G, Morvan B (1996) Ruminal methanogenesis and its alternatives. Ann Zootech 313-318.
- Stackhouse KR, Pan Y, Zhao Y, Mitloehner FM (2011) Greenhouse Gas and Alcohol Emissions from Feedlot Steers and Calves. Journal of Environmental Quality 40(3): 899-906.
- Huarte A, Cifuentes V, Gratton R, Clausse A (2010) Correlation of methane emissions with cattle population in Argentine Pampas. Atmospheric Environment 44(23): 2780-2786.
- Shibata M, Terada T (2010) Factors Affecting Methane Production and Mitigation in Ruminants. Animal Science Journal 81(1): 2-10.
- Brask M, Lund P, Weisbjerg MR, Hellwing ALF, Poulsen M, et al. (2013) Methane Production and Digestion of Different Physical Forms of Rapeseed as Fat Supplements in Dairy Cows. J of Dairy Science 96(4): 2356-2365.
- Steinfeld H, Wassenaar T (2007) The role of livestock production in carbon and nitrogen cycles. Annu Rev Environ Resour 32: 271-294.
- Kristensen T, Mogensen L, Knudsen MT, Hermansen JE (2011) Effect of Production System and Farming Strategy on Greenhouse Gas Emissions from Commercial Dairy Farms in a Life Cycle Approach. Livestock Science 140(1-3): 136-148.
- Bunglavan SJ (2014) Methanogenesis and recent techniques for mitigation of methanogenesis in ruminants. Journal of Livestock Science 5: 35-48.
- Zinder SH (1993) Physiological ecology of methanogens. In: Methanogenesis (Ferry JG, ed) Chapman of Hall, New York, London, 128-206.
- Cramer B, Franke D (2005) Indications for an active petroleum system in the Laptev Sea, NE Siberia. Journal of Petroleum Geology 28(4): 369-384.
- Katz BJ (2011) Microbial processes and natural gas accumulations. The Open Geology Journal 5(1).
- Kietäväinen R, Purkamo L (2015) The origin, source, and cycling of methane in deep crystalline rock biosphere. Frontiers in microbiology 6: 725.
- Thauer RK (1998) Biochemistry of Methanogenesis: a Tribute to Marjory Stephenson. Microbiology 144: 2377–2406.
- Conrad R (1999) Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS microbiology Ecology 28(3): 193-202.
- Rjwilmsi (2020) Methanogenesis.
- Finazzo C, Harmer J, Bauer C, Jaun B, Duin EC, et al. (2003) Coenzyme B induced coordination of coenzyme M via its thiol group to Ni (I) of F430 in active methyl-coenzyme M reductase. Journal of the American Chemical Society 125(17): 4988-4989.
- Hungate RE (1967) Hydrogen as an intermediate in the rumen fermentation. Archiv für Mikrobiologie 59(1-3): 158-164.
- Hungate RE, Smith W, Bauchop T, Yu I, Rabinowitz JC (1970) Formate as an intermediate in the bovine rumen fermentation. Journal of Bacteriology 102(2): 389-397.
- Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, et al. (2015) Global dispersion and local diversification of the methane seep microbiome. Proceedings of the National Academy of Sciences 112(13): 4015-4020.
- Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B (2010) The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465(7298): 606-608.
- Krüger M, Meyerdierks A, Glöckner FO, Amann R, Widdel F, et al. (2003) A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426(6968): 878-881.
- Faulet BM, Niamke S, Gonnety JT, Kouame LP (2006) Purification and biochemical properties of anew thermostable xylanase from symbiotic fungus, Termitomyces. African J of Biotechnology 5(3): 273-282.
- Griggs DJ, Noguer M (2002) Climate change 2001: the scientific basis. Contribution of working group I to the third assessment report of the intergovernmental panel on climate change. Weather 57(8): 267-269.
- Metcalf WW, Griffin BM, Cicchillo R M, Gao J, Janga SC, et al. (2012) Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science 337(6098): 1104-1107.
- Yusuf O, Raifu Z, Noor Z, Ahmad H (2012) Green House Gas Emissions: Quantifying Methane Emission from Livestock. Am J Engg. & Applied Sci 5(1): 1-8.
- Patra AK (2012) Enteric methane mitigation technologies for ruminant livestock: a synthesis of current research and future directions. Environmental Monitoring and Assessment 184(4): 1929-1952.
- Patterson JA, Hespell RB (1979) Trimethylamine and methylamine as growth substrates for rumen bacteria andMethanosarcina barkeri. Current Microbiology 3(2): 79-83.
- McAllister TA, Okine EK, Mathison GW, Cheng KJ (1996) Dietary environmental and microbiological aspects of methane production in ruminants. Can J Anim Sci 76: 231-243.
- Miller TL, Wolin MJ, de Macario EC, Macario AJ (1982) Isolation of Methanobrevibacter smithii from human feces. Applied and environmental microbiology 43(1): 227-232.
- Martin C, Morgavi DP, Moreau D (2010) Methane mitigation in ruminants: from microbe to the farm scale. Animal 4: 351-365.
- Lassey KR (2008) Livestock Methane Emission and Its Perspective in the Global Methane Cycle. Australian Journal of Experimental Agriculture 48(2): 114-118.
- Keppler F, Hamilton JT, Braß M, Röckmann T (2006) Methane emissions from terrestrial plants under aerobic conditions. Nature 439(7073): 187-191.
- Le Mer J, Roger P (2001) Production, oxidation, emission and consumption of methane by soils: a review. European journal of soil biology 37(1): 25-50.
- Purkamo L, Bomberg M, Kietäväinen R, Salavirta H, Nyyssönen M, et al. (2016) Microbial co-occurrence patterns in deep Precambrian bedrock fracture fluids. Biogeosciences. 13 (10): 3091–3108.
- Newberry CJ, Webster G, Cragg BA, Parkes RJ, Weightman AJ, Fry JC (2004) Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environmental Microbiology 6(3): 274-287.
- Kotelnikova S (2002) Microbial production and oxidation of methane in deep subsurface. Earth-Science Reviews 58(3-4): 367-395.
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe R (1979) Methanogens: reevaluation of a unique biological group. Microbiological reviews 43(2): 260.
- Ashby KD, Casey TA, Rasmussen MA, Petrich JW (2001) Steady-state and time resolved spectroscopy of F420 extracted from methanogen cell and its utility as a marker for fecal contamination. J Agri Food Chemistry 49(3): 1123-1127.
- Sirohi SK, Pandey N, Singh B, Puniya AK (2010) Rumen methanogens: a review. Indian J Microbiol 50(3): 253-262.
- Hook SE, Wright ADG, McBride BW (2010) Methanogens: methane producers of the rumen and mitigation strategies. Archaea, 2010.
- Baker SK (1999) Rumen methanogens, and inhibition of methanogenesis. Australian J of Agri Res 50(8): 1293-1298.
- Steudler PA, Bowden RD, Melillo JM, Aber JD (1989) Influence of nitrogen fertilization on methane uptake in temperate forest soils. Nature 341(6240): 314-316.
- Sharp R, Ziemer CJ, Stern MD, Stahl DA (1998) Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiology Ecology 26(1): 71-78.
- Johnson KA, Johnson DE (1995) Methane emissions from cattle. Journal of animal science 73(8): 2483-2492.
- Mc Ginn SM, Beauchemin KA, Coates T, Colombatto D (2004) Methane emissions from beef cattle: Effects of monensin, sunflower oil, enzymes, yeast, and fumaric acid. J Anim Sci 82: 3346–3356.
- Palut MPJ, Canziani OF (2007) Contribution of working group II to the fourth assessment report of the intergovernmental panel on climate change. (Vol. 4). Cambridge University Press.
- Nair A (2015) After Freedom Park, waste to light up Gandhinagar in Bengaluru. The Economic Times.
- Young A (1992) Land fill gas migration. DoE Report CWM039A+B/92.
- BBC (2005) Methane as sign of life.
- European Space Agency (ESA) (2004) Methane in Martian Atmosphere.
- Howell S (2017) Cassini-Huygens: Exploring Saturn's System.
- Knapton S (2014) Life on Mars: NASA finds first hint of alien life. The Telegraph.
- New Scientist (NS) (2017) Atmospheric methane.
- National Geographic (NG) (2004) Methane as sign of life.
- Northon K (2017) NASA Missions Provide New Insights into 'Ocean Worlds'. NASA.
-
Getabalew M, Alemneh T, Bzuneh E. Review on Methanogenesis and its Role. World J Agri & Soil Sci. 6(2): 2020. WJASS. MS.ID.000632.
-
Acetoclastic, Anthropogenic, Hydrogenotropic, Methane, Methanogenesis, Methylotropic
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






