 Research Article
Research Article
Military Shooting Range Xenobiotic Bacteria Consortiain Situ Biodegradation, Kachia, Kaduna, Nigeria
Ayodele A Otaiku1* and Isyaku A Alhaji2
1Department of Environmental Management, Nigerian Defence Academy, Nigeria
2Department of Microbiology, Nigerian Defence Academy, Nigeria
Ayodele A Otaiku, Doctoral student, Department ofEnvironmental Management, Faculty of Arts and Science, Nigerian DefenceAcademy, Kaduna, Nigeria.
Received Date: December 30, 2019; Published Date: January 21, 2020
Abstract
24.95 square kilometers Nigerian military shooting range established in 1965 was studied (June-August 2015 and February-March 2016)for in situ bacteria consortia impacts on xenobiotic biodegradation using polymerase chain reaction amplification of 16srRNA gene. The opticaldensity reading of biodegrading activity of each bacterial isolate on explosives broth with 1% exposure mineral salt medium (MSM) was evaluated.Mixed bacterial consortia (MBC) observed high bioremediation potential (68.62%) compared to single isolate in decreasing order were Bacillussubtilis, 55.1%; Enterobacter spp, 54.80%; Escherichia coli, 54.1%; Arcobacter spp, 43% Klebsiella pneumonia 42.7%, Lysini bacillus 37.8% andAchromobacter spp, 31.5% respectively attributed to the synergistic effect between the catabolic enzymes in the eight bacteria isolates. Anova testfor total explosive contents shows a significant difference for all locations in both dry and wet Seasons (P<0.05) according to the Duncan multiplerange test. All bacteria isolate reduced heavy metals better than the control in decreasing order were Bacillus subtilis>Enterobacter spp>Escherichiacoli>Arcobacter spp>Klebsiella pneumonia >Lysinibacillus >Achromobacter spp. MBC observed high bioremediation potential (68.62%) comparedto single isolate. MBC has the highest and prominent percentages reduction on heavy metal pollutants as a consequence of microbial ecology,bacterial strains having plasmid linked degradation or chromosomal genes, reduction ability, substrate specificity influenced microbial remediationof the explosives xenobiotic. Habitat filtering principle and resource ratio theory explains microbial completion and genetic information of morethan one organism is necessary to degrade the complex mixtures pollutants as expressed in the MBC biodegradability (Figure 1).
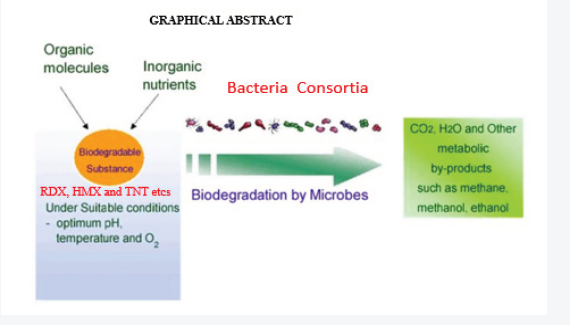
Keywords: Shooting range; 16srRNA; Polymerase chain reaction; Bioremediation; Xenobiotic; Bacterial consortia; Microbial competition;Explosives; Enzymes; Heavy metals; Biodegradation.
Introduction
The manufacture use and disposal of explosives as well as thedemilitarization of military facilities can result in environmentalcontamination. The three most widely used explosives are2,4,6-trinitrotoluene (TNT), hexahydro-1,3,5-trinitro-1,3,5-triazine
(RDX) and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX).Technological advancements in microbial bioremediation includethe conversion of slow biodegradation process into a rapid bymeans of several modern methods including microbial fuel cells(MFCs) [1,2], bioreactors [3], biofilms [4] and the use of microbialconsortia for the degradation of recalcitrant organic compounds[5]. In ex-situ bioremediation technologies for explosives such ascomposting [6-8]. Biological based remediation is an economicaland ecological compatible approach to detoxify areas contaminatedwith xenobiotics [4,9,10]. The reductive reactions are the basisof several treatment processes for the bioremediation of TNTcontaminated soils [11-14]. However, TNT is resistant to oxidativemicrobial degradation and only low mineralization rates havebeen sporadically reported with bacterial consortia. Instead ofoxidation, many bacteria catalyse the reduction of one or twonitro-groups of TNT to mono amino di nitrotoluenes (ADNT) anddiamino nitrotoluenes (DANT). Another pathway is mediated byaddition of one or two hydride ions to the aromatic ring, resultingin the formation of Meisenheimer-complexes (adducts of aromaticnitro-compounds with a nucleophile) often accompanied byrelease of nitrite. The electron transfer is catalyzed by differenttypes of cytoplasmatic nitroreductases [15,16]. Also, precursorsand metabolites of TNT are classified as very toxic, carcinogenicand mutagenic [17-19]. Typical contaminated sites may containup to 10 g/kg TNT in soils and up to 100 mg/l in water. TNTand its metabolites exhibit a high toxic and mutagenic potentialon both prokaryotes and eukaryotes [20-23]. TNT and some ofits degradation products have a high persistence, toxicity andmutagenicity [20]. The objective of the research was to isolatesthe bacteria organisms in the military shooting range to ascertaintheir biodegrading capabilities on explosives (RDX, TNT, HMX) andheavy metals contaminants and the impacts of the mixed bacterialconsortia (MBC) biodegradability.
Microbial bioremediation
In general, single microbial species hardly degrades anyorganic substrate in isolation [24] and works well in community.Interactions between microbes in the community ensure theexchange of genetic information between microbial speciesconferring resistance, tolerance and chemical-degrading ability.These processes are further subjected to the varying environmentalconditions influencing the growth pattern of microbes [25]Microbiological removal of metal ions from the environment is anew biotechnique [26] and the most cost-effective approach inmitigation of elemental pollution. It is imperative to know thatmicrobes in any case cannot degrade the metal ions and are onlyable to transform metallic ions from higher to lower oxidationstates to stabilize them [27]. Successful microbial bioremediationis achieved when microbes interact within their niche, under themost favourable conditions [28]. Biostimulation (enhancing theactivity of native microbes); bioaugmentation (increasing theviable microbial counts); bioaccumulation (storage of toxic ornontoxic elements by the microbes) [29,30]; biosorption (removalof elements from the environment through adsorption) [31] wheremicrobial transformation of metal ions involve the action of specificenzymes for oxidation, reduction, methylation, dealkylation andprecipitation [32] and the use of biofilms [4] are some of theexamples of recent advancements in microbial bioremediationtechniques.
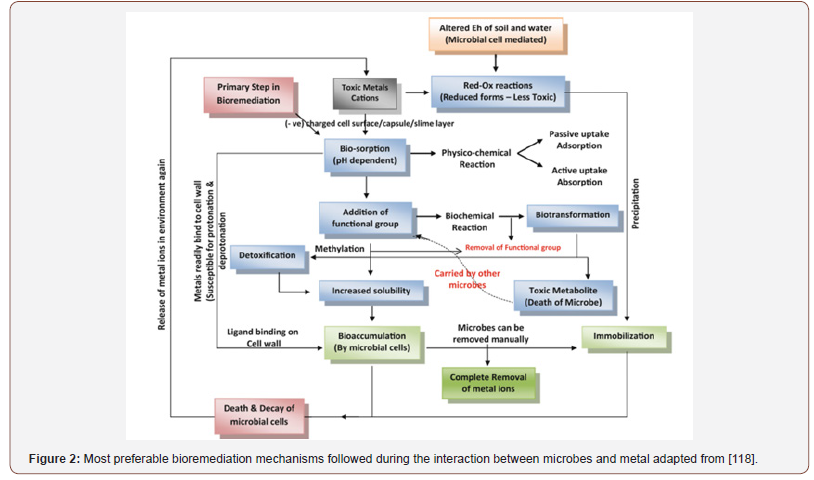
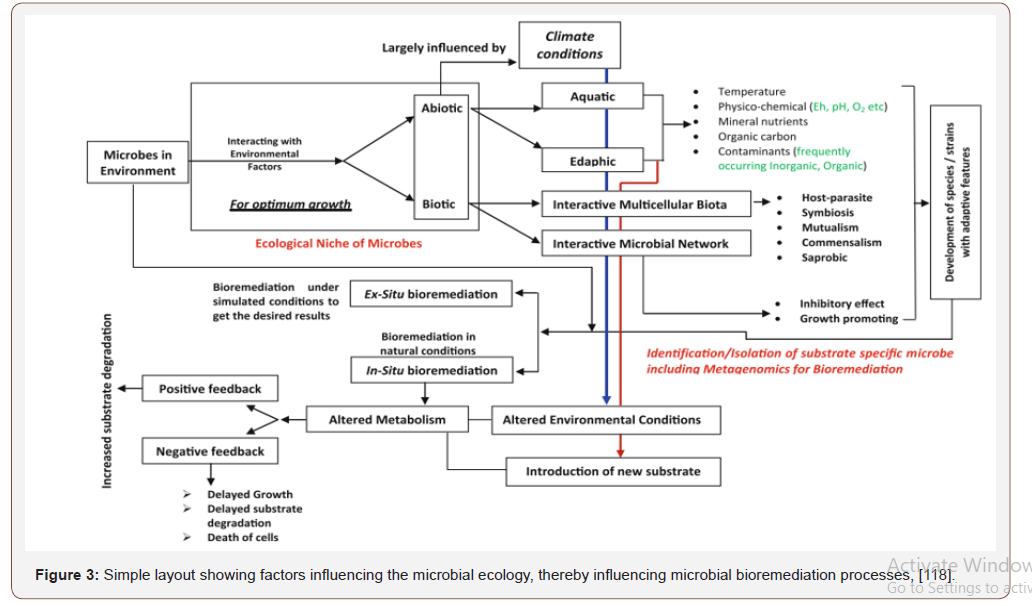
Higher microbial counts and diversity is directly associatedwith the higher content of organic matter in soil [33]. Generally,organisms rely upon associations with their neighbours for thesustenance of life [34]. Microbes also interact with other organismswithin their surroundings for organic carbon, energy and shelterto metabolize, detoxify and accumulate metals mostly in cell walllike any other nutrient element. Microbes releasing chelatingagents and acids altering physicochemical properties such as redoxpotential (Eh) of surroundings [35] bring substantial changesby increasing the bioavailability of metal ions [36]. Microbeshave diversified conditions for their growth with ability to adaptto the changing environmental conditions [37]. Advancementsin microbial remediation techniques and studies on plasmidencodedbiochemical information and genetic engineering helpedin designing new strains of known bacterial species for targetedbioremediation, e.g., recombinant Escherichia coli expressingmetallothionein gene (Neurospora crasa) for cadmiun uptake,which was more rapid than the gene donor microbial species[38]. First step in the interaction between microbial cell and metalions [32] followed by the physical adsorption, ion exchange andcomplexation, (Figure 2). Microbes have inherent ability of firstrate to get adapted to the changing environment though they havecertain limitations. In-depth understanding of microbial ecology isrequired for the improvement in the microbiological action and topredict the successful bioremediation process [25]. Figure 3 showsthe environmental challenges that microbes face in their life.
Three types of factors affecting the microbial processes
• Physicochemical characteristics of environment or theabiotic factors,
• Biological factors or biotic factors and
• Climatic conditions whereby physicochemical andclimatic conditions are among the major factors affectingthe metabolic rates in microbes.
Materials and Methods
Study site
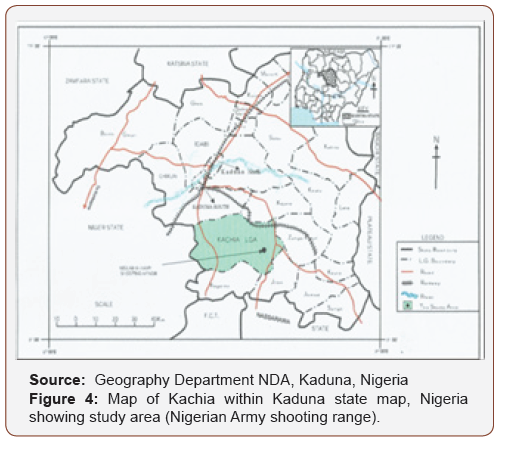
The study was conducted in the permanent military shooting/training range located at 5 km east of Kachia town in Kaduna state,north central Nigeria. The shooting range was established in 1965and it covers an area of about 24.95 square km that lies betweenlongitudes 90 55’ N and 70 58’ E, with an elevation of 732 m abovesea level and the topography is undulating and the vegetation isguinea savannah, Nigeria ecology. The area where the munitions/explosive are fired (the impact area) is a valley consisting of aboutfour large rocks, where the fired munitions/explosives land andexplode during military training. Five military exercises involvingthe deployment of explosives are carried out annually by theNigerian Defence Academy (NDA) Kaduna, Nigerian Air force (NAF)Kaduna, Nigerian Army School of Infantry (NASI) Jaji, Armed ForcesCommand and Staff College (AFCSC) Jaji and Nigerian Army Schoolof Artillery (NASA) Kachia, Kaduna, Nigeria, (Figure 5).
Sampling points
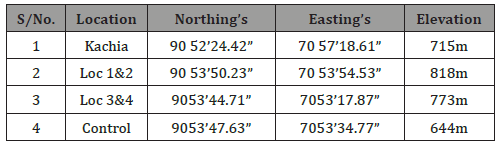
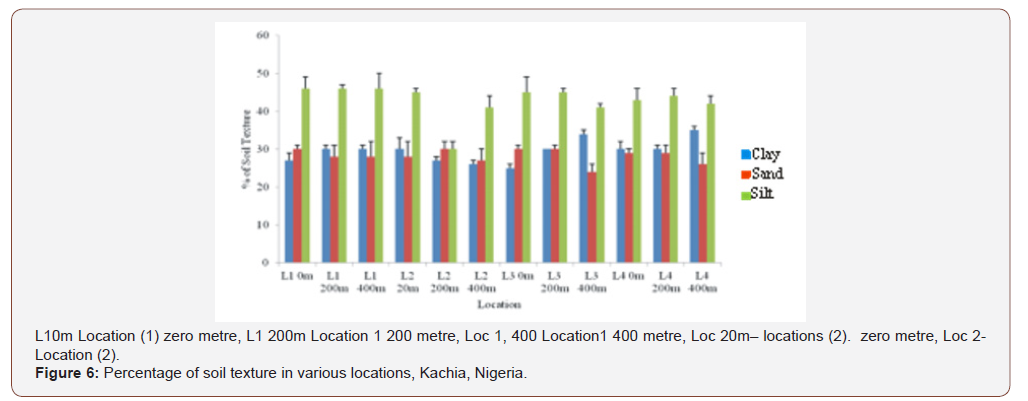
Four sampling points selected for the study are locations 1, 2, 3and 4, Table 1. Locations 1 and 2 are the twin smallest rocks closestto the road. While locations 3 and 4 are much larger rocks heavilyimpacted by munitions/explosives between locations 1 and 2 is aflat ground where rain run offs flow through to the stream in the study area (map with global positioning system [GPS] co-ordinates).Locations 1 and 2 approximately 200m from the Plateau top, wherethe small arms are fired such as Kalashinokov, FN, Greenade, GPMG,SMG and Pistols. The soil in locations 1 and 2 is made of 50% siltand a flat ground with shrubs and drainage that flow through tothe farmlands near the sites. Location 3 is approximately 9000maway from the Plateau top lies between 90 53’ 44.71” Northingsand 70 53’ 17.87” Eastings. The impact area of locations 3 and 4 aremainly largely rocks containing high concentration of explosivesdue to the extensive use of bombardment by the artillery weapons,155mm nortwizer, and other heavy weapons while location 4 isahead of location 3 is about 10,000m from the Plateau top whereheavy weapons are fired. The distance between locations 3 and 4was covered with various shrubs and two major streams, Figure 4,Plates 1 and 2, respectively.
Sampling technique and Soil treatments
Soil sampling: Sampling was done during both dry and wetseason, (Table 6 in Appendix 2). Four locations located withinNASA shooting/training range Kachia were earmarked as samplingsites for this study using soil iron auger.10 gram of soil sample (0-30cm in depth) with diameter of 9 cm were collected from 3 differentpoints within a location and harmonized to form a compositesample at various locations of the sites. All sample collected weresieved using a 63 (106m) mesh size laboratory sieve and thenstored in black labelled polythene bags until for analyses. Samplesfor microbial analyses Table 3 were kept in a cool box refrigeratedwith ice pack to retain the original microbial activities (Table 5 inAppendix 1).
Soil sample pre-treatment: Sampling points were treated inthe laboratory before digestion commence. 10 grams of the soilsample was weighed into a clean dried beaker and put into an ovenat about 100°C for one hour. The soil sample was then ground in aporcelain mortar with pestle and sieved through 250 μg mesh sizeto obtain a homogenous sample.
The soil sample was stored in sterilized polyellylere bags,label and kept for next stage of pre-treatment. This procedure wasrepeated for all the collected soil samples. The ground soil sampleswere used for analyzing heavy metal and explosives content for soilsamples [39].
Table 1: Global positioning system (GPS) recorded in degrees, minutesand seconds (DMS) of sampling locations..
Digestion of soil sample for heavy metals and explosivesanalysis: 1 g of pre-treated soil sample was accurately weighedand placed in a 100 cm3 beaker. Then, 4 cm3 of 70% of HCLO4 and2 cm3 of concentrated HNO3 were added to the sample and theresultant mixture was put in the oven at 100 0C overnight. A whiteash was formed which was dissolved in 2 cm3 of 1m HCL the digestwas filtered into 50 cm3 standard volumetric flask. The beakerwas then rinsed with small portions of double distilled water andquantitatively transferred into the flask. The resultant solution wasmade up to the mark with double distilled water, transferred into aclear dried plastic sample bottle, labelled and kept for heavy metaland explosives analysis. This procedure was repeated for all thesamples collected at this site. A blank was prepared using the sameprocedure excluding the soil sample.
Heavy metals analysis principle
The analysis was done using Atomic AbsorptionSpectrophotometer (AAS) at the laboratory of Geological SurveyKakuri- Schimadzu AAG50 model [40]. The digested soil sampleswere analyzed for heavy metals. Heavy metals analyzed includearsenic, boron, cadmium, chromium, cobalt, copper, iron, lead,magnesium manganese, nickel and zinc using AAS, accordingto APHA (1998) [41]. The technique makes use of absorptionspectrometry to assess the concentration of analyzed in a sampleby measuring the absorbance against a known concentration. Itrelies on the Bear Lambert Law.
Explosive analysis
The analysis was carried out at National Research Institutefor Chemical Technology (NARICT), Zaria, Nigeria. The vialscontaining the AcN (Acetonitrite) soil extracts were placed into GasChromatography (GC) auto sampler trays that were continuouslyrefrigerated by circulating 0°C glycol/water through the trays, theextracts were analyzed by GC using a micro-electron capture detector(GC-μECD). Results were obtained on HP-6890 GC equipped with amicro cell Ni63 detector at 280°C according to the general procedureoutlined, in EPA SW-846 method 8095 [42]. Direct injection μL ofsoil extract was made into a purged packed inlet port, at 250°C, thatwas equipped with a deactivated Restek Uniiner. Primary analysiswas conducted on a 6 m x 0.32 mm ID-fused - silica column, witha 1.5-um film thickness of 5% (Phenyc) - methyl - siloxane (RTX-5 from Restek). The GC oven was temperature programmed as follows; 100°C for two minutes, 10°C minutes ramp to 260°C, twominutehold. The carrier gas was helium at 10 ml/minute (linearvelocity approximately 90 cm/second). The ECD make-up gas wasnitrogen flowing at 40 ml/minute. If a peak was observed in theretention window for a specific signature compound, the extract wasreanalyzed on a confirmation column, 6 mx 0.53 mm ID having a 0.1mm film thickness of 50% cyanopropylmethyl - 50% phenylmethylpolysiloxane(RTX - 225 from Restek). If analyze concentrationswere taken from the determination on the primary column, unlessthey appear to be coalition with another compound, in, such casesreported concentration were taken from the determination of theconfirmation column.
Screening for explosive/munitions degrading organisms
Nutrient Agar: Nutrient Agar (Antec/USA) by dissolving 28g ofthe Agar in one litre of distilled water in a conical flask. The conicalflaks with the media was auto daved at 121°C at 15 Pressure PerSquare (PSI) for 15 minutes. Cooled to 400C before pouring ordispensing if a sterile petri dishes.
Mackonkey Agar: Mackonkey Agar was prepared by dissolving49.53g of dehydrated medium in 1000ml distilled water in a conicalflask. It was heated to dissolve the medium completely. The mediumwas sterilized by autoclaving at 15 1bs pressure (121°C) for 15minutes. Cooled to 45-50°C, mixed well before pouring into sterilepetri plates.
Potato Dextrote Agar (PDA): Thirty-nine (39)g of dehydratedmedium suspended in 1000 ml distilled water was heatedto dissolve the medium completely. It was then sterilized byautoclaving at 15 1bs pressure (121°C) for 16 minutes. Cooled tenPercent (10%) of tartanic acid was added and mixed well to achievepH 3.5 before dispensing into petri plates. The media were spreadwith the specimen soon after solidification of the media. Plateswere incubated at 25-30°C in an inverted position (agar slide up)with increased humidity. Cultures were examined weekly for fungalgrowth and were held for 4-6 weeks.
Basal Salts Medium (BSM): Composition of what is BSAmedium for, isolation and growth of bacteria degrading explosives.The ingredients in the order listed were added to 800 ml of tap waterwhile stirring. Until the last ingredient was dissolved completely.The final medium was filtered through fine filter papers. Agar (2%)was added and sterilized in an autoclave at 121°C and 15 pressureper square (PSI) inch for 15 minutes. The normal pH is 6.9 to 7.0.Bacteria microbes like Bacillus spp have efficient biodegradativepatways for xenobiotics [43].
Isolation of possible explosives degrading bacteria
Thirty (30) gram samples of soil from selected positivescreening tests was placed into flasks and amended with onepercent (1%) co-metabolite (sodium acetate) and 5 ml of distilledwater.
Flasks were plugged and incubate at 28°C. After one week ofincubation samples of the acetate primed sterile water were platedon to basal salts medium supplemented with 15g/l yeast extractcannot solidify medium nutrient agar and the co-metabolite ata concentration of 20g/l and 60 g/ml nystatin to suppress fungalgrowth. Plates were incubated at 28°C for two weeks. Representativecolonies were picked from each plate and transferred to liquidculture media containing 100 mg/L TNT and incubated for 10 daysat 28°C on a gyrorotary shaker at 150 rpm. Bacteria Isolates wereidentified by Blog identification (7 series of biochemical tests inliquid for metabolic characterization) and confirmed by referenceto Bergey’s manual of systematic bacteriology.
Serial dilution: The sterile dilution blanks were marked in thefollowing manner. 100 ml dilution blank was 102 and 9 ml tubessequentially were 103, 104,10-6. One gram of soil sample wasweighed from each sample located and added to the 12-2 dilutionblank and vigorously shaken for at least one full minute with thecap securely tightened. All the 10-2 dilution was allowed to sitfor a short period. The one ml from this dilution was asepticallytransferred to the 10-3 dilution was again transferred to the 10-4dilution. The procedure was done to 10-5 and 10-6. A flask ofnutrient agar from the 45° C water bath was especially poundedinto each petri plates for that set. 15 ml was poured enough to coverthe bottom of the plate and mixed with the one ml inoculum in theplate. Each set was gently swirled on the bench so that the inoculumgets thoroughly mixed with the ager. All the plates were allowed tostand without moving so that the agar solified and set completely.The plates were inverted and stacked into pipette carnisters andplaced in the incubator or at room temperature until after 48 hoursthe same procedures were applied for MacKonkey agar and PDA.
Isolation of possible heavy metals tolerant bacteria
The soil samples solution were inoculated into nutrient agarand mackonkey agar plates containing different concentrationsof the heavy metals in their salt form such as Lead chromate (vi)(PbCrO4), Nickel chloride (NiCl3), Cadmium nitrate (CdNO3). Zincsulphate (ZnSO4), Manganese (iv) Oxide (MnO2), Cobalt aluminates(CoAl2O3), Chromium chloride (CrCl3), Iron arsenosulphide (FeAs5)and Arsenic Trichloride (AsC3). The plates were incubated at 30°Cfor 24-48 hours. The bacteria were isolated and subculture intonutrient agar plates to obtain pure culture of each tolerant strain.
Pure culture of bacteria
Streak method was used to subculture the bacteria isolate frombasal salt medium (BSM) with 15g/l agar containing 100 mg/lTNT and co-metabolite at a concentration of 20 g/l. An inoculatingloop was used to pick each colony gently from the Basal salt agarplates. The colony was re-streaked gently onto the surface of a freshnutrient agar plate. The bacteria plates (Table 3) were incubated atroom temperature on a laboratory bench [44].
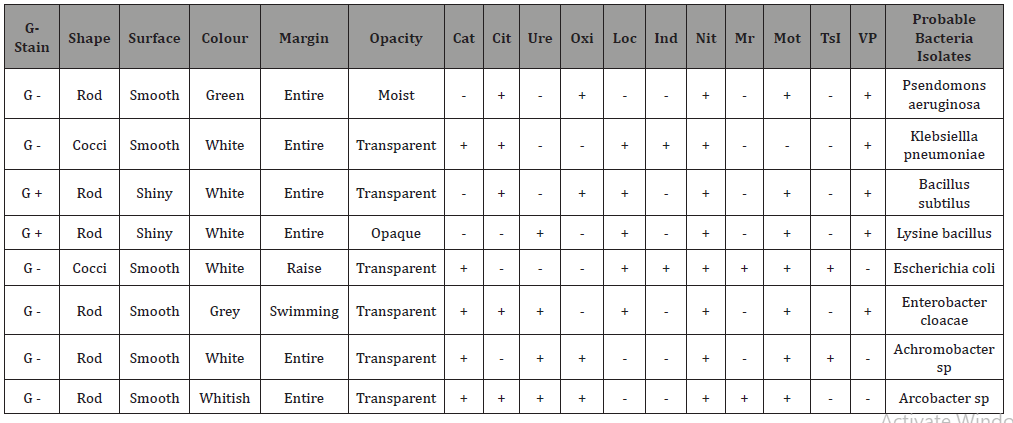
Characterization of bacterial isolates gram staining: Thinsmear of colony from the petri dish containing the bacterial culturewas prepared. The smears were air dried and heat fixed using sliderack. The smears were stained with crystal violet for 30 seconds,each slide was washed with distilled water for a few seconds using wash bottles and then each smear was treated with gram’s iodinesolution for 60 seconds. The iodine solution washed off with 95%ethyl alcohol. Ethyl alcohol was added drop by drop, until no morecolour flows from the smears. The gram positive bacteria werenot affected while all gram negative bacteria were completelydecolorized. The slides were washed with distilled water, allowedto be drained and treated with safranin smears for 30 seconds andwas washed with distilled water and blot dry with absorbent paperand the stained slides was allowed to air dry. The bacterial thatretained the primary stain (crystal violet) were gram positive andthose that do not retain the primary stain were gram-negative, [44].The slides were examined microscopically using oil immersionobjective. Bacterial colonies were identified by gram’s staining andbiochemical characterization according to Bargy’s Manual.
morphology of bacteria isolates: Isolated colonies ofeach bacteria were absorbed and culturally morphologically basedon their superficial forms (circular, filamentous and irregular)elevation (flat, convex and umbonate), Margin (Lobate, convex orirregular), and shape spiral, rod or cocci using hand magnifyinglens.
Biochemical characterization of bacteria catalase testprinciple
The enzyme catalase is present in some bacteria whichbreakdown hydrogen peroxide to water and oxygen. The principlewas used for the detection of catalase enzyme in a bacterial isolated.Few colonies of the test organism were picked with a platinum loopfrom nutrient and Mackonkey Agar as the case may be and thenapplied in a drop of 10% hydrogen peroxide on a clean sterile slide.The production of gas bubble from the culture indicated positivereaction.
Citrate test: The simmons create agar medium contains the pHindicator bronollymol blue and medium citrate as the sole sourcesof carbon. Citrate is an enzyme found in some bacteria. Citrateis breakdown by citrate to oxaloactic acid and acetic acid. Thebreakdown is indicated in the medium by the change of colour fromgreen to blue. However, the removal of citrate from the mediumby bacteria that can utilize citrate creates an alkaline condition inthe medium and indicator changes to its alkaline colour blue. Thetest organism were inoculated into simmon’s citrate agar slantsand incubated at room temperature for 24 hours. Colour changefrom green to blue indicated a positive test while no colour changeindicated a negative result.
Coagulase test: Coagulase is an enzyme aureus in staphylococciaureus that causes the conversion of soluble fibrinogen intoinsoluble fibrin. The reaction is different from the normal clothingmechanism exhibited in blood clothing. Gram positive staphylococcithat produced catalase positive reaction were further tested forcoagulase. 0.5ml suspension of a 24 hours broth culture of thebacteria was mixed with 1ml of anticoagulant containing bloodplasma in a test tube. This was incubated at room temperatureand positive coagulase bacteria revealed the formation of fibrincloths in the test-tubes. Positive coagulase is a characteristic ofstaphylococcus aurens aura [45].
Mortality indole orinthine agar
Mortility can be determined from Brownian motion (small,random movement) exhibited by the bacteria in the medium.Trptophanase is oxidized by an enzyme trypotophanase in somebacteria to produce indole pyruvate react with Kovaes, reagent(dimellhycamino-benzaldehyde) to form resounded (cherry redcompound) and water. Bacteria with Ornithine decarboxylasewill decorboxylate the amino acideornithine into purescine. Themedium which contains bromcyresol purple with turn yellow afterthe enzyme inedecarboxylateornithine.The amino acid ornithineinto putrescine. The medium which contains bomcresol purplewill turn yellow after the enzymedecarboxylate ornithine. Mortilityindole ornithine (M10) medium was used to determine motility andindole production. Ten (10) ml of the medium was dispensed intoa test-tube and autoclaved. Each pure colony of the unidentifiedorganism was stabbed into the test tube using a sterilized wireloop and incubated at room temperature for 24 hours. Motilitywas observed as hazy diffuse spreading growth (Swarm) along thestab unlike non motles, restricted to the stab line. The presence ofornithine decarboxylase is observed when the purple colour of themedium changes to yellow [46]. For indole production 2-3 dropsof Kovac’s reagents (Dimellylamino-benzylaldehyde) was addedto the medium in the test tube. A red precipitate at the top of theinterface indicated a positive reaction of indole production. ForOrnithine decarboxylase reaction [47].
Methyl red test: Some bacteria produces large amount oforganic acids such as formic, lactic acid and succinic acid. Theseorganic acids react with methyl red resulting in red coloration ofthe medium. The test was conducted to detect the production ofsufficient acid by fermentation of glucose so that pH dropped to 4.5.The test organism were inoculated in glucose phosphate broth andincubated at room temperature for 2-5 days. Five drops of 0.04%solution of methyl red were added and mixed well. A bright redcolour indicated a positive result while a yellow colour indicated anegative result.
Oxidase test: Certain bacteria contain a terminal cytchrome Cand its associate oxidase found in their respiratory chain. Bacteriawhich contains such a chain can oxidize chemicals such as Kovac’sOxidase reagent (1% tetramethy l-p-phencle-nediamine dihydrochloride). Electrons are transferred from the chloride). Electronsare transferred from the reagent to cytochrome and hence, throughthe oxidase to oxygen. When oxidized, the reagent develops anintense violet colour. A drop of 1% solution of oxidase reagent wasadded on a piece of filter paper. Few colonies of the test organismwere rubbed onto it. The production of a deep purple colour within10 seconds indicated a possible reaction.
TSI test: Triple sugar iron agar contains glucose, lactose,sucrose, sodium thiosulphate and a ferrous. The fermentationof any of the sugar results in a yellow colorations of the mediumbutt or slant. Hydrogen sulphide is produced from the reduction of amino acids such as cysteine and methiomine. Hydrogen sulphidewhen produced reacts with the metal salt (ferrous sulphide) to forma visible black. Ferrous sulphide precipitates. If gas is produced,this would give risk to crack in the butt of the medium due to theproduction of oxygen from the organism stab into the butt of themedium. Agar slants were prepared in test-tubes. Each pure colonyof the unidentified bacteria was picked with a sterilized wire loopand stabbed into the butt of the test tub. Colonies were serpetinelystreaked on the slope before stabbing into each butt and allowed touncubate at 30°C for 24 hours. When only glucose was fermented,the slant was red (alkaline) and butt turned yellow. If lactose orsucrose was fermented as much acid was produced that both theslant and butt remained yellow (acid). Gas production resulted ina cracks or the medium was pushed upward. H2S production wasconfirmed by the change of the medium colour to black along thestreak line.
Nitrate reduction: The nitrate test is used to determinebacterium that can reduce nitrate. The medium consist ofpotassium nitrate which when reduced turn the medium fromyellow to red. The test organism was inoculated in 5 ml mediumcontaining potassium nitrate, popatone and distilled water whichwere incubated at 200m temperature for 96 hours. 0.1 ml testregent which consist of equal volumes of 0.8% sulphanilic acid and0.5% naphylamine in 5N acetic acid was mixed before use. A redcolour developing of nitrate hence the ability of the test organismto reduce nitrate to nitrate.
Urease test: The urea medium contains the pH indicator phenolred and urea. Urea is a major organic waste product of proteindigestion in most bacteria. Urease is a hydrolytic enzyme in somebacteria which catalyze urea into carbon dioxide and ammonia. Theammonia liberated would dissolve to create an alkaline mediumand the indicator would change to its alkaline colour red. The abilityof an organism to produce urease enzyme was evaluated by thistest: The test organism were separately inoculated on the entireslope of Christensen’s medium which contains urea and phenol andindicator. It was incubated at room temperature and examined after4 hours overnight incubation development of red colour indicatedproduction of urea enzymes by test organism.
Isolates of explosive degrading bacteria from kachiafiring range
Eight sets of 250 ml flasks were prepared for the work. Thenutrient broth was sterilized in an autoclave at 1210 for 15minutes.120 ml of nutrient broth was incorporated with tenmillimetermineral salt and 1% explosive concentration into eachof the nine flasks. The first flask was left as blank while second,third, fourth, fifth; sixth, seventh and eighth flasks were inoculatedwith each of the isolated bacteria. The ninth flask was inoculatedwith a mixed culture consortium of the seven bacteria isolates.It was incubated for 14 days at room temperature. The growthof the bacteria was measured by taking the optical density (O.D)using a spectrophotometer at 600 nm from 0 hour to 14 days atregular interval of 2 days against a blank containing broth with 1%explosive concentration [48].
Bacteria isolates to various concentrations of someheavy metals
The isolated bacteria were each inoculated into a nutrient brothcontaining different concentration of heavy metal, 0.5mg/ml, 1mg/ml, 2 mg/ml, 4 mg/ml, 6 mg/ml and 8 mg/ml respectively for Pb,Ni, Cd, Zn, Mn, Co, Cr, Cu and As. The experiment was left for 24hours and the inhibitory concentrations were measured using aspectrophotometer at an absorbance of 600nm against a nutrientbroth (blank) containing the same amount of heavy metals.
Isolation of genomic DNA from bacteria
Bacteria with proven bioremediation capacity (also efficientstrains of these bacteria) was selected and the DNA isolated. DNA ofbacteria strains isolated was extracted from 1ml of bacteria culture,the culture was pelleted by centrifuging at 12,000 rpm for 2 minutes,the pellet was treated with lyses solution and proteinase K andincubated at 600C for 30 minutes. Nucleic acid were precipitatedwith isopropanol by centrifuging at 10,000 rpm for 10 minutes,washed with 1ml of a 70% (v/v) ethanol solution and dissolved in0.1ml of a TE buffer (Figures 5&6). The purity and quantity of DNAwere examined by recording its agarosegel electrophoresis [48,49].
Bioremediation of heavy metals in soil
The soil samples solution was sterilized using an autoclave at120°C for 15 minutes. 100 ml of the sterilized soil solution waspoured into fifteen conical flasks. The first flask was not inoculatedwith bacteria (control), the second, third, fourth, fifth, sixth,seventh, eighth, ninth, tenth, eleventh, twelfth and thirteenth flaskswere each inoculated with Acbrombacter spp; Bacillus subtilis,Lysinibacillus spp, Arcobacter spp, E. coli, Enterobacter spp andEscherichia spp respectively. The experiment was left to stand for24 hours after which the concentration of Cadmium, Chromium,Nickel, Zinc, Cobalt, Iron, Lead, Cupper and Arsenic were measuredusing Atomic Absorption Spectrophotometer (AAS).
Bioremediation of explosive in soil
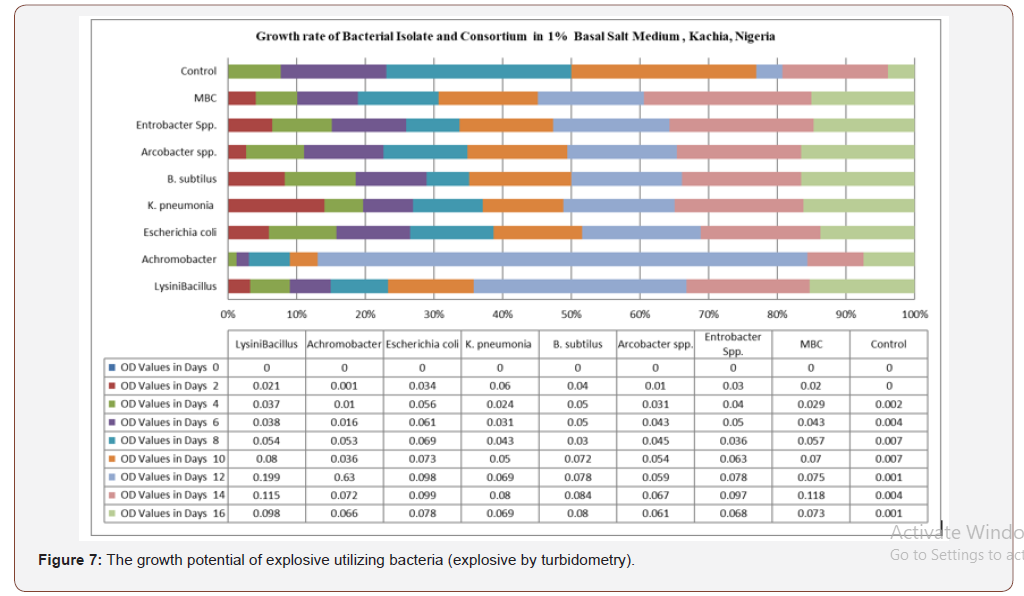
Nutrient broth and soil solution were added in a ratio 4:1.8 setof 250 ml flask were set for the work. The broth and soil solutionwere sterilized in an autoclave at 121°C for 15 minutes. 120 ml ofnutrient broth and 30 ml of sterilized solid solution were addedinto each of the fifteen flasks. The first flask was left uninoculated,but the second flask up to the eleventh flasks was inoculated withthe isolated bacteria and fungi from explosive/munitions whichare Achromobacter spp., Bacillus subtilis. Lysinibacillus spp.,Enterobacter spp, Escherichia coli. The experiment set-up wasleft to stand for 14 days and the micro remediation was measuredby taking the optical density (O.D) reading at 595 minutes from 0hours-14 days at regular intervals of 2 days against the control [50],Figures 6&7.
Polymerase chain reaction amplification of 16srRNAGene
The PCR reaction mixture containing 10X PCR buffer, 25mm, magnesium chloride, 25 mm dNTP’s, 10pm/ul Primerconcentrations and template DNA were used for the amplification of the 16srRNA gene for each isolates. PCR conditions were optimizedusing lab net thermal cycler. The PCR program began with an initial5-min denaturation step at 94°C: 35 cycles of 94°C for 45 seconds,annealing (1 minute at 55°C), 10 minutes extension step at 72°C.All reaction mix were preserved at 4°C until it was time for analysisas reported by Kloos in 2006. The amplified 16srRNA gene of eachisolates was further characterized using gel electrophoresis.
Sequence determination of 16srRNA gene: The amplified16srRNA gene of each isolate was processed for sequencing andcharacterization. The sequencing Kit (Applied Biosystems) withthe product was analyzed with ABI prism DNA sequence (ABI).The gene sequence of each isolates obtained in this study werecompared with known 16srRNA gene sequences in the Gene Bankdatabase as described by [48].
Bacterial biodegradative activity by turbidometry: Eightsets of 250 ml flasks were set for the work. The broth was sterilizedin an autoclave at 121°C for 15 minutes. 120 ml of nutrient brothwas inoculated with 1% munitions/explosives concentrationinto each of the seven flasks. The first flask was left uninoculatedwith bacteria isolates but the second, third, fourth, fifth sixthand seventh were inoculated with the isolated bacteria whichare Enterobacter, Escherichia, Klebsiella, Arcobacter, Bacillus,Lysinibacillus, Achromobacter spp respectively. The eight flask wasinoculated with a mixed culture consortium of the seven bacteriaand was measured by taking the Optical Density (O.D) readings at595nm from 0 hours - 14 days at regular intervals of 2 days againstmineral salt medium as blank [47].
Statistical analysis
Data collected from the study were analyzed using generaldescriptive statistics, one way Analysis of variance (ANOVA) at95% probability level of significance. If significant differences werefound. Duncan’s Multiple Range (DMR) test was used to comparethe different experimental groups. Computer Software StatisticalPackage for Social Scientists (SPSS) and Microsoft-Excel were usedfor the statistical analysis.
Polymerase chain reaction (PCR) for amplification ofcatabolic genes
PCR consist of an exponential amplification of DNA fragmentand the principle is based on the mechanism of DNA replication invivo, double stranded DNA is denatured to single stranded DNA,each single strand DNA is anneal by the forward and reverse primersof known sequence and elongated using tag DNA polymerase toproduce copies of DNA template. Isolated explosive bacteria DNAwas amplified using catechol 2,3 dioxygenase gene primers. Thegenomic DNA, the primers and PCR master mix were added intoa PCR tube. The tubes were spun to collect the droplets. The tubeswere then inserted into the PCR machine while maintaining theregulation for initial denaturation (45 seconds at 94°C), annealing(1minute at 55°C) extension (1minute at 72°C) and final extension(10 minutes at 72°C). The amplified catabolic genes were resolvedin 1.5% agarose gel electrophoresis stained with ethidium bromideand viewed under ultra-violet (UV light).

Results and Discussion
Physio-chemical properties of the military firing ranges
The study area 34.22°C dry season and 27.16°C wet seasonwere recorded as an integral of the climate of the region; alsowithin similar work by Jenkins [51] 27-35°C on characterization ofexplosives contamination at military firing ranges .The explosivesvalues obtained within Federal Ministry of Environment,Nigeria (FMENV) limit of <40 μg kg-1 with soil pH of 6-8 valuesuitable microbial bioremediation [52]. The high value of electricconductivity EC recorded during the wet season (Table 4, Appendix1) similar to finding of Oaikhena in 2015 and by Duniya [53]respectively. The value of (EC) is above the FMENV and WHO [54-57] permitted limits of 500/μ5/cm. The low values of total dissolvedsolutes for both wet and dry season were [58] within the value of2000 mg/l acceptable by FMENV. Which is contrary to the finding ofOaikhena in 2015, whose values were within 2400 - 5900 mg/l. andis in agreement with Usman [50] and Duniya [53] the high valuesof total dissolve solid (TDS) would be due to the deposit of organicand inorganic matters necessitated by the military activities, Figure4. Xenobiotic analyzed during dry season are higher than those inwet season as indicators of the soil pollution detrimental to bothhuman and the environment (Table 6, Appendix 2). The soil hadlow clay percentage which facilitates increase ventilation (Figure 4).Increased ventilation has a direct impact on microbial growth whichcan enhance the biodegradation of explosives. The water holdingcapacity of the contaminated soil samples under investigation wasobserved during this study (59.56) is in the ranger suggested foroptimal bioremediation [52]. From the result in (Table 5 Appendix2) it was observed that the concentration of heavy metals variessignificantly (P>0.05) between the concentration of toxic metals inboth dry and wet season samples which invariably will affects thebacteria biodegradation of the pollutants of concern as affirmed onTNT degrading bacteria.
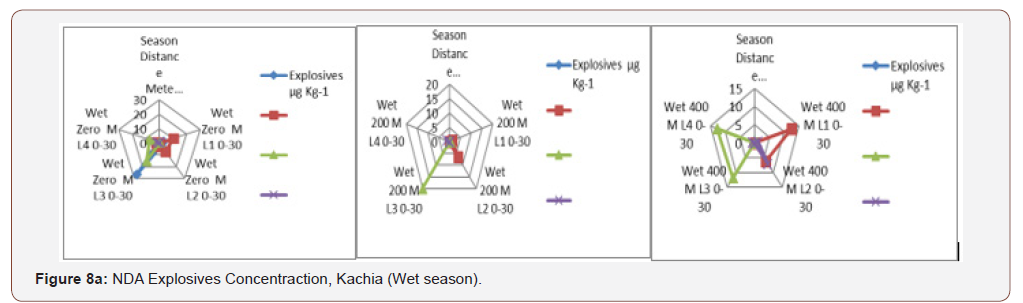
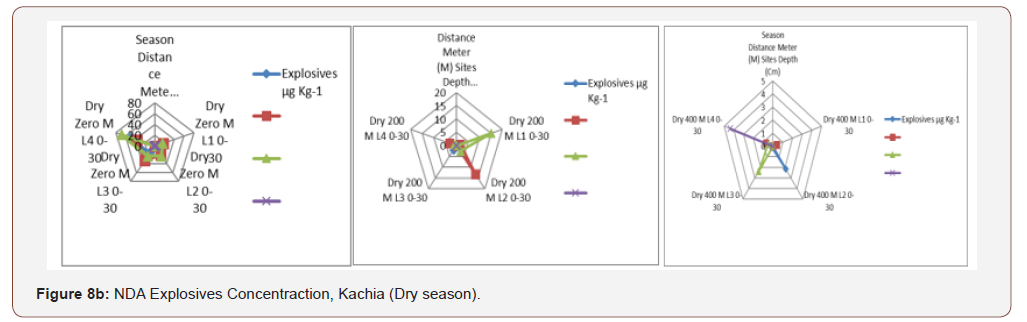
Concentration of the toxic metals was also observed to behigher in the contaminated site than the uncontaminated sitebecause of the activities of military activities affirmed by similarwork. The average concentration of the metals was higher duringthe dry season than the wet season, as a results of metals washedas runoff into the drain as a result of rainfall, Table 6. Whereas, thelower level concentration of heavy metals found in the dry seasoncould be attributed to high temperature which may have constantly account for increase and decrease in the rate of radiation in dry andwet season [59]. The result shows that lead (Pb) with (27.01) mg/kg) in location 1 which was significantly higher (P>0.05) than othermetal is probably a recalcitrant metal to micro-remediation (Table6, Appendix 2). WHO has recommended >10μg/l lead as safepermissible level in the drinking water. In zero metre, TNT, RDX andHMX were consistently found at the highest values of 49.39 mg/kg,68.19 mg/kg and 35.67 mg/kg, respectively.
An impact area and beyond within the hand grenade range, a105 mm howitzer firing point and the heavy artillery and mortarrange that had been analyzed by GC-ECD. TNT concentrationsranged from 0.11 to 49.39 mg/kg in dry season and 0.12 to 26.6mg/kg in wet season. HMX concentrations ranged from 0.69 to35.67 mg/kg in dry season and 1.43 to 28.21 during wet season,then RDX concentrations ranged from 0.49 to 68.19 mg/kg in dryseason and 0.38 to 19.9 mg/kg during wet season, while, PETNconcentration ranged from 0.39 to 4.38 mg/kg in dry season and0.17 to 7.12 mg/kg during wet season similar to the report [50].Among all the concentration of explosives both in dry and wetseason RDX recorded the highest concentration of 68.19 mg/kgimplies that the Kachia shooting range is polluted with explosivesmost especially the RDX and agreed by EPA according to Lachance[22].
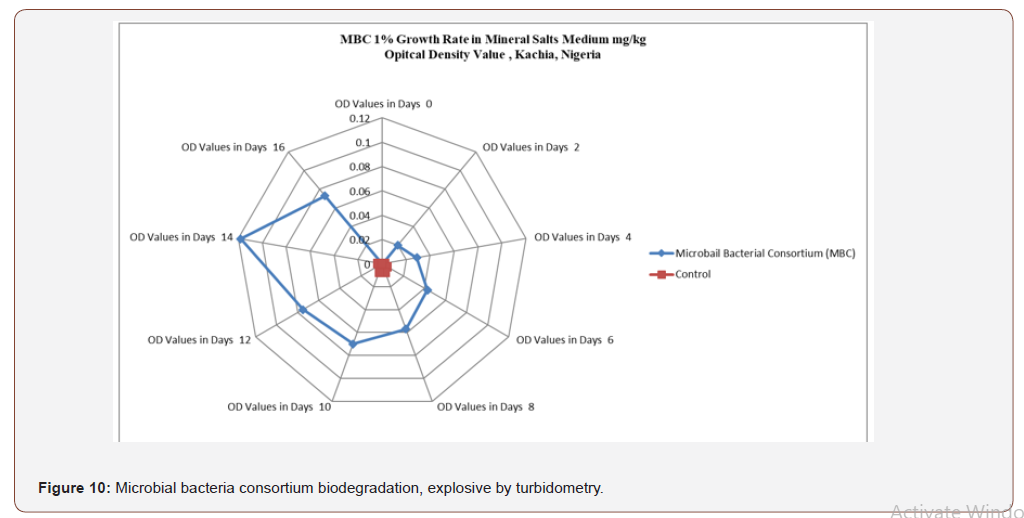
Bacteria Consortium Explosives Degradation Bacteriaconsortium biodegradation: Explosive by Turbidometry
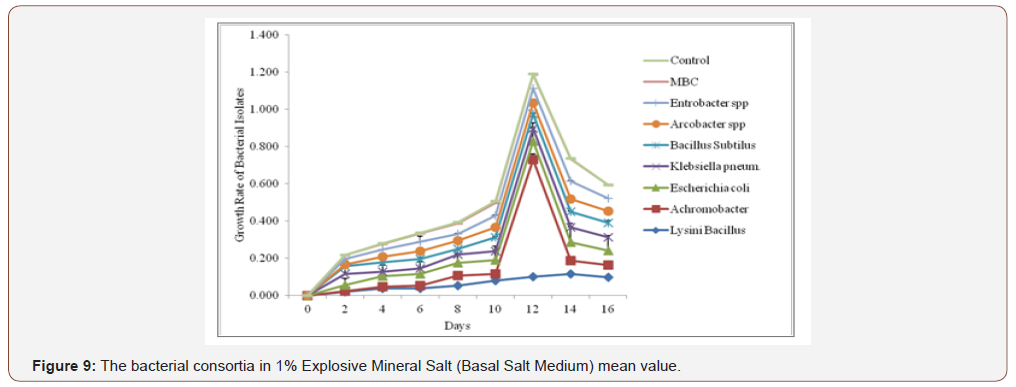
Figure 7 shows the optical density reading of biodegradingactivity of each bacterial isolate on explosives broth with 1%exposure mineral salt medium (MSM). The results revealed thatthe bacteria Lysinibacillus spp, Achromocter spp, E. coli, Klebsiellapneumonia, Bacillus subtilis, Arcobacter spp and Enterobacter spp,had the ability to degrade explosives. The result of analysis showsthat there is a significant difference in the overall growth rate readingat 595 nm for 14 days of incubation, Figure 8. The overall resultsalso indicated that growth rate increase significantly from the 8thto 14th day Achromobacter and Arcobacter had Lag, exponential,stationary and death phases. Lysinibacillus, E.coli and microbialbacteria consortium (MBC) growth rate observed was exponential,stationary and death phases while Bacillus subtilus, KlebsiellaPneumonia and Enterobacter spp. exhibited stationary, exponentialand death phases. The test on the degrading activity of isolates onexplosive show that bacteria genus Lysinibacillus, Achromobacterspp, E.coli, Klebsiella Pneumonia, Bacillus subtilis, Arcobacter spp,Enterobacter spp and MBC were potent degraders of explosive withMBC (Table 2) > Lysinibacillus > E.coli > Enterobacter spp > Bacillussubtilis > Klebsiella pneumomia > Achromobacter > Arcobacterspp with optical density values of 0.182, 0.118, 0.099, 0.097, 0.084,0.080, 0.072 and 0.067 respectively . Growth of bacteria between0-2 days was slow. Meanwhile, the number of bacterial speciesthat had the peak in day 12 were Lysinibacillus, E. coli and Bacillussubtilis respectively (Figure 9). Anova test for total explosivecontents shows a significant difference for all locations in both dryand wet Seasons (P<0.05) according to the Duncan multiple rangetest, (Table 6, Appendix 2).
Bacteria heavy metals biotransformation andmineralization
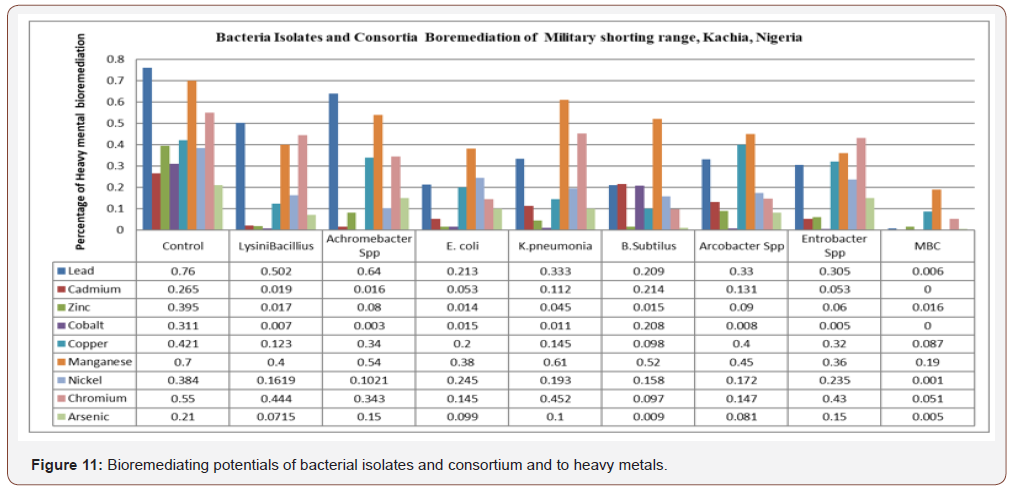
Bioremediating potentials of bacterial isolates andconsortium and to heavy metals: In Figure 11 certainbacteria isolate with different percentage reduction of heavymetals was observed when compared with control and valuesobtained. Lead concentration was reduced by Lysinibacillus to25.8%, Achromobacter spp to 12%, E. coli to 54.7%, Klebsiellapneumonia to 42.7%, Bacillus subtilis to 55.1%, Arcobacter sppto 43% Enterobacter spp to 65.5% and MBC to 75.4%. Cadmiumwas reduced by Lysinibacillus to 24.6%, Achromobacter spp to24.86%, E.coli to 21% K. pneumonia to 15.3%, B. subtillis to 100%.Arcobacter spp to 13.14%, Enterobacter spp to 21.2% and MBC to100%. Zinc was reduced by Lysinibacillus to 37.8%, Achromobacterspp to 31.5%, E. colito 38.1%, K. pneumonia to 35%, B. subtillisto 38%, Arcobacter spp to 30.5%, Enterobacter spp to 35.5%and MBC to 37.9%. Cobalt was reduced by L. bacillus to 30.4%,Achromobacter spp to 30.8%, E. coli to 29.6% K. pneumonia to 30%B. subtilis to 100%, Arcobacter spp to 30.3%, Enterobacter spp to30.6 and MBC to 100% (Figures 11&12). Copper was reduced by L. bacillus to 29.8%, Achromobacter spp to 8.1%, E.coli to 22%, K.pneumonia to 27.6%, B. subtilis to 32.3%, Arcobacter spp to 2.1%,Entrobacter spp to 10.1% and MBC to 32.4%. Manganese wasreduced by L. bacillus to 30%, Acromobacter spp to 16%, E.colito 32%, K. pneumonia to 9%,B. subtilis to 18%, Arcobacter spp to25%, Entrobacter spp to 34% and MBC to 51%. Nickel was reducedby L. bacillus to 22.21%, Achromobacter spp to 28.19%, E. coli to14.1%, K. pneumonia to 19.1% B. subtilis to 22.6%, Arcobacter sppto 21.19%, Enterobacter spp to 14.9% and MBC to 38.3%.
Table 2: The growth rate (optical density values) of microbial bacterial consortium..
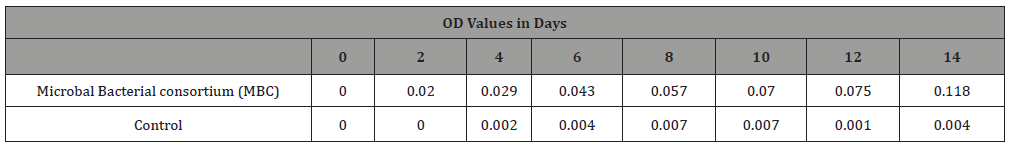
Table 3: Gram Stain Result, Morphological and Biochemical Characterization of the different microbial Isolates from the NASA Shooting Range KachiaKaduna..
Chromium was reduced by L bacillus to 10.6%, Achromobacterspp to 20.7%, E. coli to 40.5%, K. pneumonia to 9.8%, B. subtilisto 45.3%, Arcobacter spp to 40.3%, enterobacter spp to 12%and MBC to 49.3%. Arsenic was reduced by L bacillus to 13.85%,Achromobacter to 6%, > E. coli to 11.1%, K.pneumonia 11% B.subtilis to 20.1%, Arcobacter spp to 12.9% Enterobacter spp to5.96% and MBC to 20.5%. All bacteria isolate reduced heavy metalsbetter than the control. The best isolates for heavy metal reductionin decreasing order were Bacillus subtilis>Enterobacter spp>E. coli >Arcobacter spp> Klebsiella Pneumonia > Lysinibacillus>Achromobacter spp. In the overall, the mixed culture consortiumhas the highest and prominent percentages reduction of eachon every heavy metal analyzed in this study. Anova test for totalexplosive contents shows a significant difference for all locations inboth dry and wet Seasons (P<0.05) according to Duncan multiplerange test, Table 6, Appendix 2. Mixed bacterial consortium (MBC)observed high bioremediation potential (68.62%) compared tosingle isolate, Figure 8 whose bioremediation potential observedin decreasing order were Bacillus subtillis, Enterobacter spp, E.coli, Arcobacter spp, Klebsiella pneumonia, Lysini bacillus andAchromobacter spp in the following order as (55.1%), (54.80%),(54.1%), (43%) (42.7%), (37.8%) and, (31.5%) respectively. MBCshowed high percentage degradation of explosive which might beattributed to the synergistic effect between the catabolic enzymesin the eight bacteria isolates. These findings are similar to the resultobtained by Duniya [53] whose research reported biodegradationrates at 97% by MBC.
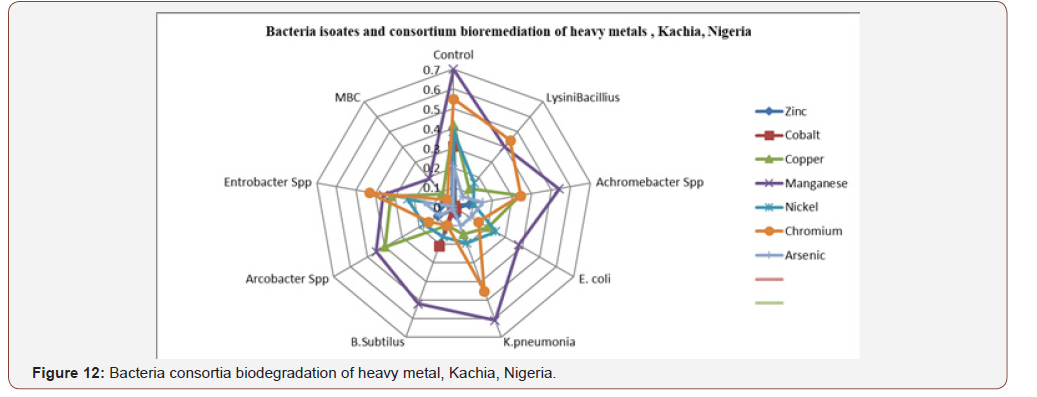
In these results, Figures 11&12 Achromobacter spp andAcrobacter spp experienced lag, exponential, stationary and deathphases which are main four phases of growth contradict with thefinding of Ojima in 2004, by Usman [53] in 2012 and Oaikhena in2015 who reports that bacteria isolates did not exhibit lag phase ofgrowth. Lysinibacillus, E.coli and MBC growth rate observed wasexponential, stationary and death phase while Bacillus subtilis,Klebsiella Pneumonia and Entcrobacter spp exhibited stationary,exponential and death phases, result similar to Usman [53] in 2012,reported and similar to Figure 12 result. The efficacy of a desired insitucatalytic activity (biodegradation) depends first on its presencein the target site. One key enzyme may not be there, or it may preexistin the site but not be manifested. Alternatively, it can be hostedby just a very minor part of the whole microbial population, so thatits factual expression in the site might not be significant [60] andallows for bio augmentation microbes inclusion for bioremediation.
Significance of microbial bacteria consortium
Generally, organisms rely upon associations with theirneighbours for the sustenance of life [34]. Microbes also interactwith other organisms within their surroundings for organic carbon,energy and shelter to metabolize, detoxify and accumulate metalsmostly in cell wall like any other nutrient element. Mixed microbialcommunities (Figure 9) have the most powerful biodegradativepotential because the genetic information of more than oneorganism is necessary to degrade the complex mixtures of organiccompounds present in contaminated areas as reported by Fritsche[61] explained the impacts of MBC of the site xenobiotics.
Microbes releasing chelating agents and acids alteringphysicochemical properties (Table 4, Figure 13, Appendix 1) suchas redox potential (Eh) of surroundings [35] bring substantialchanges by increasing the bioavailability of metal ions [36], Figure4. Higher microbial counts and diversity is directly associated withthe higher content of organic matter in soil [33] narrated in Table 5,Appendix 1.The consequence is the application of biosimulants onthe treated and post treated xenobiotics site as reported by Muter in2012 biostimulate the capacity of bacteria to degrade TNT becauseseveral inherent characters of microbes that affect the substratedegradation [62]. Xenobiotics requires complex multispeciesinteractive networks for biodegradability and reported in similarwork by Pandey and Jain [63] with plasmid-encoded genes providespecificity for substrates because organohalide respiratorybacteria thrive only in consortia, and their isolation and culture isvery difficult. [64] reported that the optimum growth conditionsof microbes are unpredictable. Initiation of microbial degradationof xenobiotics depends on the toxic pressure exerted by thecontaminants on the microbes that induce enzymatic modifications[63], Figure 14, Appendix 3.
Klebsiella pneumomia bacteria inoculum degrade TNT by up to90 % after 14 days incubation as reported by Muter in 2012 thisis a function of Nitroreductases which are ubiquitous in bacteriaand localized in the cytoplasm where they are constitutivelyexpressed as Type I nitroreductases. This is expressed in gramnegativebacteria, such as Escherichia coli [65], Enterobactercloacae [66,67], Klebsiella spp. Kim and Song [16] they catalysethe pyridine nucleotide dependent reduction of nitro-aromaticcompounds which are related to the old yellow enzyme (OYE) ofyeast [16,68,69]. Gram-negative bacteria are the best candidates forthe microbial incorporation in self-cleaning explosive formulationssince they are more tolerant to TNT than Gram-positive bacteria[70].
Escherichia coli bacteria has been genetic engineering onplasmid-encoded biochemical information recombinant expressingmetallothionein gene for improved Cd uptake in reports byPazirandeh [38] and abHan Han [32] which could have implicationsfor cadmium reduced by E.coli to 21% compared to MBC to 100%,Figure 11 biodegradation expressed in the interaction betweenmicrobial cell and metal ions,as reported by Han and Gu, 2010.Several studies have reported the anaerobic degradation of RDXand HMX by pure bacterial cultures failed in aerobic conditionexcept anaerobic condition under a microbial consortium likeEnterobacteriaceae, Citrobocter freundii. Bacteria may possessreduction mechanisms that are not coupled to respiration, butinstead are thought to impart metal resistance. For example,reduction of Cr (VI) to Cr (III) under aerobic condition [71].
Microbial competition and biodegradability
Stepwise denitration of RDX involves a nitrate reductase whichis a ubiquitous enzyme possessed by a diverse group of bacteria,especially denitrifying bacteria [72] which affirm the role of bacteriaconsortium during bioremediation. Degradation of RDX throughdenitration and ring-cleavage involves the microbial P450 systemwhich was shown to be able to degrade RDX under both aerobicand anaerobic conditions [73]. The main degradation pathwaysdescribed involve either a sequential biotic reduction of the nitrofunctional groups followed by abiotic ring-cleavage [74] or a directdenitration followed by hydration and subsequent ring-cleavage[73] also reported the role of micobes consortia in bioremediation,(Figure 3&14).
Reduction of metals can occur through dissimilatory metalreduction [75], where bacteria utilize metals as terminal electronacceptors for anaerobic respiration are the mechanisms ofbioremediation of the sites xenobiotics .Environments with a highnutrient complexity Figure 4, Plate 1 and Plate 2 respectively,containing multiple resources or niches, can reduce selection forcompetition [76], particularly if each species is limited by a differentresource (Resource Ratio Theory) [77,78] explained in Figures 8& 9 by Lysini bacillus, E.coli and MBC growth rate observed wasexponential, stationary and death phase while Bacillus subtilis,Klebsiella Pneumonia and Entcrobacter spp exhibited stationary,exponential and death phases, result similar to Usman [50], thatelaborated the microbial competition, Figure 14, Appendix 3.Thetwo main resources necessary for microbial survival are nutrientsand space from the pollutants site. Nutrients essential for microbialgrowth and metabolic functions include: carbon, nitrogen,phosphorus, sulphur, hydrogen, calcium, iron, and other metals [79-82]. A third and less commonly considered resource is geneticmaterial. DNA is used as a nutrient source, but it may also provideits host with beneficial traits, enhancing its ability to survive andadapt [83].
Empirical data from this and other microbiome studiesindicate that, in agreement with the ‘habitat filtering’ principle,species with similar resource requirements tend to live in similarareas of the body [84-86] which may explain local competition.Another possibility is that, as weaker strains are outcompeted inthe soil, diversity is reduced. And because high diversity isolatescompetitors from each other through buffer zones [87,88] novelwarfare may be enhanced between the remaining strains asthe buffer zones disappear. [89] have shown using a theoreticalmodel, that this can lead to ecologically stable equilibria whereindifferent species neutralise all produced antibiotics, and diversity ismaintained. Apart from ecological conditions, different strains canremain mixed in space if they depend on each other for growth andsurvival [87,90-92]. The presence of neighbouring colonies alsoalters the competitive behaviour of many species of soil bacteria[76,93,94]. However, this may occur in a number of different waysas narrated in Figure 14, Appendix 3 xenobiotic biodegradation.
Three ecologically stable outcomes of competition are wellaccepted
• the less competitive strains go extinct while othersdominate the community [93,94]
• strains continue to coexist by occupying differentmetabolic niches, where each specialises on a differentresource type, or
• strains separate into different spatial niches or patches.Habitat filtering’ principle using Escherichia coli (speciescommunity member) can metabolically shift fromfermentation to respiration when oxygen is present,generating high growth rates but low yield, allowingthem to absorb nutrients faster than their competitors[97-100].
Of the best-studied systems involving the interplay betweenthese two competitive mechanisms, cooperation that allows moreaccess to nutrients, and cheating that saves the cost but relies onthe presence of co-operators is the production of iron-chelatingsiderophores [101-105] and of quorum sensing (QS) molecules thatcoordinate the expression and production of exofactors [106,107]enzymes, rhamnolipids allow cells to swim to new areas or pushcompetitors away [671,108] adhesins bind to surfaces and preventdisplacement by invaders [69] extracellular polysaccharides(EPS) can smother and starve competitors, while also pushingclone-mates into nutrient-rich environments [109-111], E. colicells produce surfactants and EPS that inhibit biofilm formationin Staphylococcus aureus and Pseudomonas aeruginosa [112,113].Bacillus subtilis produces enzymes that degrade QS molecules inVibrio cholerae, which is subsequently unable to form biofilms[112,114].
Conclusion
This study explained the munition synergistic interactionsenvironments (Table 6, Appendix 2) with a high nutrientcomplexity in the military shooting range, Kachia (Figures 9&10),containing multiple resources or niches, can reduce selection forcompetition particularly if each species is limited by a differentresource (Resource Ratio Theory) must rely on evolving orengineering metabolic codependence in a mixed culture (bacteria)for survival and adaptation for bioremediation to occur (Figure13 & Table 5). Microbiome studies indicate that, in agreementwith the ‘habitat filtering’ principle, species with similar resourcerequirements tend to live in similar areas of the body whichexplain local competition with the microbial bacteria consortium,Figure 14, Appendix 3. Apart from ecological conditions, differentstrains can remain mixed in space if they depend on each other forgrowth and survival based on the as narrated in Table 2, Figure9. Competition can become neutralised through a reduction ininteraction strength, potentially leading to symbiotic relationshipsand productive communities [115-119]. The dynamics of stabilityand diversity, then, strongly depend on environmental conditions,and the nature of the competitive phenotypes for collaborationwould lead to more accurate and informed predictions on thenature of interactions in microbial communities. The ability tomake such predictions can have many important implications in themanagement and design of microbial communities for xenobioticbiodegradation and understanding of microbial competition formicrobial bioengineering narrated (microbial competition andsustained biodegradability, Appendix 3, Figure 14).
Acknowledgment
None.
Conflicts of Interest
The authors have not declared any conflict of interests
References
- Rabaey K, Verstraete W (2005) Microbial fuel cells: novel biotechnology for energy generation. Trends Biotechnol 23(6): 291-298.
- Moris JM, Jin S (2012) Enhanced biodegradation of hydrocarbon contaminated sediments using microbial fuel cells. J Hazard Mater 30 (213-214): 474-477.
- Robles-Gonzalez IV, Fava F, Poggi Varaldo HM (2008) A review on slurry bioreactors for bioremediation of soils and sediments. Microb Cell Factor 7(5): 1-16.
- Singh SN, Tripathi RD (2006) Environmental bioremediation technologies. Springer Berlin
- Torres-Bojorges AX, Buitro NG (2012) Biodegradation of nonylphenols using nitrifying sludge, 4- chlorophenol adapted consortia and activated sludge in liquid and solid phases. Environ Technol 33 (13-15): 1727-1737.
- Williams RT, Ziegenfuss PS, Sisk WE (1992) Composting of explosives and propellant contaminated soils under thermophilic and mesophilic conditions. J Ind Microbiol 9: 137-144.
- Griest WH, Stewart AJ, Tyndall RL, Caton JE, Ho C Het al. (1993) Chemical and toxicological testing of composted explosives-contaminated soil. Environmental Toxicology and Chemistry 12(6): 1105-1116.
- Jarvis AS, McFarland VA, Honeycutt ME (1998) Assessment of the effectiveness of composting for the reduction of toxicity and mutagenicity of explosive-contaminated soil. Ecotoxicology Environmental Safety 39(2): 131-135.
- Alvarez PJ, Illman WA (2005) Bioremediation and Natural Attenuation. Process Fundamentals and Mathematical Models pp.609.
- Crawford RL, Lynch J, Crawford DL (2005) Bioremediation: principles and applications. Cambridge University Press.
- Breitung J, Bruns-Nagel D, Steinbach K, Kaminski L, Gemsa D (1996). Bioremediation of 2,4,6-trinitrotoluene- contaminated soils by two different aerated compost systems. Appl Microbiol Biotechnol 44(6): 795-800.
- Lenke H, Achtnich C, Knackmus HJ (2000) Perspectives of bioelimination of poly nitro aromatic compounds. In: Spain JC, Hughes JB and Knackmus HJ (eds) Biodegradation of nitroaromatic compounds and explosives. CRC Press Boca Raton pp. 91-126.
- Fuller ME, Hatzinger PB, Rungmakol D, Schuster RL, Steffan RJ (2004) Enhancing the attenuation of explosives in surface soils at military facilities: combined sorption and biodegradation. Environ Tox Chem 23(2): 313-324.
- Lewis TA, Newcombe DA, Crawford RL (2004) Bioremediation of soils contaminated with explosives. J Environ Manag 70(4): 291-307.
- Pak JW, Knoke KL, Noguera DR, Fox BG, Chambliss GH (2000) Transformation of 2,4,6- trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C. Appl Environ Microbiol 66(11): 4742-4750.
- Kim HJ, Song HG (2005) Purification and characterization of NAD(P)H dependent nitroreductase I from Klebsiella sp. C1 and enzymatic transformation of 2,4,6- trinitrotoluene. Appl Microbiol Biotechnol 68(6): 766-773.
- Schneider K, Oltmanns J, Hassauer M, Schuhmacher US (2000) Determination of test values for selected armament-specific contaminants. In: Order of the Environment Agency, understates Berlin, Germany, 298: 76-251.
- Haarck T, Erdinger L, Boche G (2001) Mutagenicity in Salmonella typhimurium TA98 and TA 100 of nitroso and respective hydroxylamine compounds. Mutat Res 491: 183-193.
- Ribeiro EN, Da Silva FT, De Paiva TCB (2012) Ecotoxicological evaluation of wastewater from 2.4.6-TNT production. J Environ Sci Health A Tox Hazard Substan Environ Eng 47(2): 184-191.
- Spanggord RJ, Stewart KR, Riccio ES (1995) Mutagenicity of tetranitroazoxytoluenes: a preliminary screening in Salmonella typhimurium strains TA100 and TA100NR. Mutat Res 335(3): 207-211.
- Honeycutt ME, Jarvis AS, McFarland VA (1996) Cytotoxicity and mutagenicity of 2,4,6- trinitrotuene and its metabolites. Ecotox Environ Safety 35: 282-287.
- Lachance B, Robidoux PY, Hawari J, Ampleman G, Thiboutout S, et al. (1999) Cytotoxic and genotoxic effects of energetic compounds on bacterial and mammalian cells in vitro. Mutat Res 444(1): 25-39.
- Maeda T, Kadokami K, Ogawa HI (2006) Characterization of 2,4,6-trinitrotoluene (TNT)-metabolizing bacteria isolated from TNT-polluted soils in the Yamada Green Zone, Kitakyushu. Japan J Environ Biotechnol 6: 33-39.
- Pieper DH, Reineke W (2000) Engineering bacteria for bioremediation. Curr Opin Biotech 11(3): 262-270.
- Watanabe, K (2001) Microorganisms relevant to bioremediation. Curr Opin Biotechnol 12(3): 237-241.
- Shukla OP, Rai UN, Dubey S (2009) Involvement and interaction of microbial communities in the transformation and stabilization of chromium during the composting of tannery effluent treated biomass of Vallisneria spiralis L. Biores Technol 100(7): 2198-2203.
- Garbisu C, Alkorta I (2003) Basic concepts on heavy metal soil bioremediation. Euro J Mineral Process Environ Prot 3(1): 58-66.
- Riccardi C, Papacchini M, Mansi A, Ciervo A, Petrucca A, et al. (2005) Characterization of bacterial population coming from a soil contaminated by polycyclic aromatic hydrocarbons (PAHs) able to degrade pyrene in slurry phase. Ann Microbiol 55(2): 85-90.
- Vasudevan P, Padmavathy V, Tewari N, Dhingra SC (2001) Biosorption of heavy metal ions. J Sci Ind Res 60: 112-120.
- Tyagi M, da Fonseca MMR, de Carvalho CCCR (2011) Bioaugmentation and bio stimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 22(2): 231-241.
- Gupta R, Ahuja P, Khan S, Saxena RK, Mohapatra H (2000) Microbial bio sorbents: meeting challenges of heavy metal pollution in aqueous solution. Curr Sci India 78: 967-973.
- Han X, Gu J (2010) Sorption and transformation of toxic metals by microorganisms. In: Mitchell R and Gu J (eds) Environmental microbiology. Wiley Blackwell Pub, Hoboken, USA.
- Boopathy R (2000) Factors limiting bioremediation technologies. Bioresource Technology 74(1): 63-67.
- Badri DV, Weir TL, Vander Lelie D, Vivance JM (2009) Rhizosphere chemical dialogues: plant- microbe interactions. Curr Opin Bio technol 20(6): 642-650.
- Abou Shanab RA, Angle JS, Delorme TA, Chaney RL, van Berkum P, et al. (2003) Rhizobacterial effects on nickel extraction from soil and uptake by Alyssum murale. New Phytol 158: 219-224.
- Abou Shanab RAI, Angle JS, Chaney RL (2006) Bacterial inoculants affecting nickel uptake by Alyssum murale from low, moderate and high Ni soils. Soil Biol Biochem 38: 2882-2889.
- Naraian R, Ram S, Kaistha SD, Srivastava J (2012) Occurrence of plasmid linked multiple drug resistance in bacterial isolates of tannery effluent. Cell Mol Biol 58(1): 134-141.
- Pazirandeh M, Chrisey LA, Mauro JM, Campbell JR, Gaber BP (1995) Expression of the Neurospora crassa metallothionein gene in Escherichia coli and its effects on heavy-metal uptake. Appl Microbiol Biotechnol 43(6): 1112-1117.
- Jaiswal PC (2011) Soil, plant and water analysis Kalyani publishers, India.
- Alloway BJ (1990) Heavy Metals in Soils. John Willey and Sons Inc, New York, USA.
- Apha (1998) Standard methods for the Examination of water and wastewater. 20th
- US EPA (2000) Method 8510: Colorimetric screening procedure for RDX and HMX in soil. In Test Methods for Evaluating Solid Waste, Physical/Chemical Methods, US Environmental Protection Agency, Washington. USA,
- Mrozik A, Piotrowska-Seget Z, Labuzek S (2003) Bacterial degradation and bioremediation of Polycyclic Aromatic Hydrocarbons. Polish Journal of Environmental Studies 12(1): 15-25.
- Aneja KR (2001) Experiments in microbiology plant pathology tissue culture and Mushroom production technology. 3rd edn, New Age International Publishers, pp.192-195.
- Madigan MT, Martinko JM, Dunlap PV, Clark DP (2009) Brock Biology of Microorganism. (12th Edn), PERSON Benjamin Cummings, Boston, USA, 365: 826-909.
- Brooke JS, Annand JW, Hammer A, Dembkowski K, Shulman ST (2009) Investigation of bacterial pathogens on 70 frequently used environmental surfaces in a large urban US University J Environ Health 71(6): 17-22.
- Prescott LM, Harley JP, Klein DA (2002) microbiology (5th edn). Mcgraw Hill.
- Jyothi K, Surendra KB, Nancy CK, Kashyap A (2012) Identification and isolation of Hydrocarbon Explosives Degrading Bacteria by Molecular Characterization. Helix 2: 105-111.
- Thenmozhil R, Arumugan K, Nagasathya A, Thajuddin N, Paneer Selvam A (2013) Studies on micro remediation of used engine oil contaminated Soil Samples. Advances in Applied Science Research 4(2): 110-118.
- Usman DH, Ibrahim AM, Abdullahi S (2012) Potentials of Bacterial isolates in Bioremediation of Petroleum Refinery Wastewater. Journal of Applied phyto technology in Environmental Sanitation 1(3): 131-138.
- Jenkins TF, CL Grant, (1996) Assessment of sampling error associated with collection and analysis of soil samples at explosives contaminated sites. Cold Regions Research and Engineering Laboratory, Hanover, New Hampshire, USA.
- US EPA (2006) United States Environmental Protection Agency Quality Assurance Management staff.
- Duniya DA, Maikaje DB, Umar YA, Ponchong AW, Daniel A (2016) Molecular characterization and determination of Bioremediation potentials of some Bacteria isolated from spent oil contaminated soil mechanic workshops in Kaduna metropolis. World Applied Sciences Journal 34(6): 750-759.
- World Health Organization (2003) Iron in drinking water Background document for development of WHO guidelines drinking water quality? USA.
- World Health Organization (2003) Zinc drinking water Background document for development of WHO guidelines for drinking water quality. USA.
- World Health Organization (2004) Cadmium on Drinking Water Background document for development of WHO guidelines for drinking water quality? USA.
- World Health Organization (2004) Manganese in drinking water Background document for development of WHO guidelines drinking water quality? USA.
- Penington JC, Brannon JM (2002) Environmental Fate of Explosives. Thermochimica Acta, 384(1-2): 163-172.
- Chehnegami A, Malayeri B (2007) Removal of heavy metals by native accumulator plants. International Journal of Agriculture and Biological Science 9: 462-465.
- Sayler GS, Ripp S ( ) Field applications of genetically engineered microorganisms for bioremediation processes. Current Opinion in Biotechnology 11(11): 286-289.
- Fritsche W, Hofrichter M (2005) Aerobic degradation of recalcitrant organic compounds by microorganisms, in environmental biotechnology: Concepts and applications. HJ Jördening and J Winter (eds), Wiley VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
- Mars AE, Kasberg T, Kaschabek SR, van Agteren MH, Janssen DB, et al. (1997) Microbial degradation of chloroaromatics: use of the meta- cleavage pathway for mineralization of chlorobenzene. J Bacteriol 179(14): 4530-4537.
- Pandey G, Jain RK (2002) Bacterial chemotaxis towards environmental pollutants: role in bioremediation. Appl Environ Microbiol 68(12): 5789-5795.
- Maphosa F, Smidt H, De Vos WM, Roling WFM (2010) Microbial community and metabolite dynamics of an anoxic dechlorinating bioreactor. Environ Sci Technol 44(13): 4884-4890.
- Whiteway J, koziarz P, veall J, sandhu N, kumar p, et al. (1998) Oxygen- sensitive nitro reductases: analysis of the roles of nfs A and nfs B in development of resistance to 5- nitrofuran derivatives in Escherichia coli. J bacteriology 180(21): 5529-5539.
- Haynes, CA, Koder RL, Miller AF, Rodgers DW (2002) Structures of nitroreductase in three states. Effects of inhibitor binding and reduction. J Biol Chem 277(13): 11513-11520.
- Shin JH, Song HG (2009) Nitro-reductase II involved in 2,4,6-trinitrotoluene degradation: purification and characterization from Klebsiella sp. CI. J Microbiol 47(5): 536-541.
- Klausmeier RE, Appleton JA, DuPre ES, Tenbarge K (2001) The enzymology of trinitrotoluene reduction. Int Biodeter Biodegrad 48: 67-73.
- Williams RE, Rathbone DA, Scrutton NS, Bruce NC (2004) Biotransformation of explosives by the old yellow enzyme family of flavoproteins. Appl Environ Microbiol 70(6): 3566-3574.
- Fuller ME, Manning JF Jr (1997) Aerobic gram-positive and gram-negative bacteria exhibit differential sensitivity to and transformation of 2,4,6- trinitrotoluene (TNT). Curr Microbiol 35(2): 77-83.
- Schluter J (2015) Adhesion as a weapon in microbial competition. ISME J 9: 139-149.
- Bhushan B, Halasz A, Spain J, Thiboutot S, Ampleman G, et al. (2002) Biotransformation of Hexahydro-1,3,5- trinitro-1,3,5, -triazine catalyzed by a NAD(P)H: Nitrate oxidoreductase from Aspergillus niger. Environmental Science Technology 36(14): 3104-3108.
- Jackson RG, Raylot E, Fournier D, Hawari J, Bruce NC (2007) Exploring the biochemical properties and remediation applications of the unusual explosive degrading P450 system XplA/B. Proc Nati Acad Sci USA 104(13): 16822-16827.
- Hawari J, Halasz A (2002) Microbial degradation of explosives. In G Bitton (ed), The encyclopedia of environmental microbiology, pp. 1979-1993.
- Fernandez PM, Martorell MM, Farina JI, Figueroa L.I (2012) Removal Efficiency of Cr6+ by Indigenous Pichia sp. Isolated from Textile Factory Effluent. The Scientific World Journal. 708213 6 p.
- Kinkel LL, Schlatter DC, Xiao K, Baines AD (2014) Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. ISME J 8(2): 249-256.
- Bellucci M, Ofiţeru ID, Beneduce L, Graham DW, Head IM et al. (2015) A preliminary and qualitative study of resource ratio theory to nitrifying lab-scale bioreactors. Microbial Biotechnol 8(3): 590-603.
- Miller TE, Burns JH, Munguia P, Walters EL, Kneitel JM, et al. (2005) A critical review of twenty years use of the Resource-Ratio Theory. Am. Naturalist 165(4): 439-448.
- Barber MF, Elde NC (2015) Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet 31(11): 627-636.
- Bren A, Hart Y, Dekel E, Koster D, Alon U (2013) The last generation of bacterial growth in limiting nutrient. BMC Syst Biol 7: 27.
- Aldén L, Demoling F, Bååth E (2001) Rapid method of determining factors limiting bacterial growth in soil. Appl Environ Microbiol 67(4): 1830-1838.
- Kehl Fie TE, Skaar EP (2010) Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14(2): 218-224.
- Borgeaud S, Metzger LC, Scrignari T, Blokesch M (2015) The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347(6217): 63-67.
- Levy R Borenstein E (2013) Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci USA 110(31): 12804-12809.
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, et al. (2010) Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4(8): 962-974.
- Smillie CS (2011) Ecology drives a global network of gene exchange connecting the human microbiome Nature 480: 241-244.
- Mitri S, Xavier JB, Foster KR (2011) Social evolution in multispecies biofilms. Proc Natl Acad Sci USA 108: 10839-10846.
- Celiker H, Gore J (2012) Competition between species can stabilize public-goods cooperation within a species. Mol Syst Biol 8:
- Kelsic ED, Zhao J, Vetsigian K, Kishony R (2015) Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521(7553): 516-519.
- Momeni B, Waite AJ, Shou W (2013) Spatial self-organization favors heterotypic cooperation over cheating. Elife 2: 1-18.
- Muller MJI (2014) Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci USA 111: 1037-1042.
- Estrela S, Brown SP (2013) Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput Biol 9(12): e1003398.
- Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R (2013) Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. M Bio 4(4): e00459-13.
- Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, et al. (2015) Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA 112(35): 11054-11059.
- Hardin G (1960) The competitive exclusion principle. Science 131: 1292-1297.
- Louca S, Doebeli M (2015) Transient dynamics of competitive exclusion in microbial communities. Environ Microbiol 18(6): 1863-1874.
- MacLean RC, Gudelj I (2006) Resource competition and social conflict in experimental populations of yeast. Nature 441(7092): 498-501.
- Pfeiffer T, Schuster S, Bonhoeffer S (2001) Cooperation and competition in the evolution of ATP-producing pathways. Science 292(5516): 504-507.
- Rendueles O, Ghigo JM (2012) Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol Rev 36(5): 972-989.
- Vulic M, Kolter R (2001) Evolutionary cheating in Escherichia coli stationary phase cultures. Genetics 158(2): 519-526.
- Morris JJ (2015) Black Queen evolution: the role of leakiness in structuring microbial communities. Trends in Genetics 31(8): 475-482.
- Morris JJ, Lenski RE, Zinser ER (2012) The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3(2).
- Cordero OX, Ventouras LA, DeLong EF, Polz MF (2012) Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA 109(49): 20059-20064.
- Ghoul M, West SA, Diggle SP, Griffin AS (2014) An experimental test of whether cheating is context dependent. J Evol Biol 27(3): 551-556.
- Kümmerli R (2009) Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc R Soc Lond Biol 276: 3531-3538.
- Diggle SP, Griffin AS, Campbell GS, West SA (2007) Cooperation and conflict in quorum- sensing bacterial populations. Nature 450(7168): 411-417.
- Schuster M, Sexton DJ, Diggle SP, Greenberg EP (2013) Acyl-homoserine lactone quorum sensing: from evolution to application. Ann Rev Microbiol 67: 43-63.
- An D (2006) Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci USA 103: 3828-3833.
- Kim, W (2014) Importance of positioning for microbial evolution. Proc Natl Acad Sci USA 111: 1639-1647.
- Xavier JB, Foster KR (2007) Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA 104(3): 876-881.
- Nadell CD, Bassler BL (2011) A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA 108: 14181-14185.
- Rendueles O (2011) Screening of Escherichia coli species biodiversity reveals new biofilm-associated antiadhesion polysaccharides. MBio 2: e00043-00011.
- Valle J, Da Re S, Henry N, Fontaine T, Balestrino D (2006) Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc Natl Acad Sci. USA 103(33): 12558-12563.
- Augustine N, Kumar P, Thomas S (2010) Inhibition of Vibrio cholerae biofilm by AiiA enzyme produced from Bacillus spp. Arch Microbiol 192(12): 1019-1022.
- Griest WH, Tyndall RL, Stewart AJ, Caton JE, Vass AA, et al. (1995) Chemical characterization and toxicological testing of windrow composts from explosives-contaminated sediments. Environmental Toxicology and Chemistry 14(1): 51-59.
- Ghoul Melanie, Mitri Sara (2016) The Ecology and Evolution of Microbial Competition. Trends in Microbiol 24(10): 833-845.
- Minz D, Rosenburg E, Ron EZ (1996) Cadmium binding bacteria: screening, and characterization of new isolates and mutants. FEMS Microbiol Lett 135: 191-194.
- Srivastava J, Chandra, H Naraian R, Kalra SJS (2013) Advances in microbial bioremediation and the factors influencing the process Int J Environ Sci Technol 11(6): 1787-1800.
- Yakimov MM, Timmis KN, Golyshin PN (2007) Obligate oil-degrading marine bacteria. Curr Opin Biotechnol 18(3): 257-266.
-
Ayodele A Otaik, Isyaku A Alhaj. Military Shooting Range Xenobiotic Bacteria Consortia in Situ Biodegradation, Kachia, Kaduna,Nigeria. Sci J Biol & Life Sci. 1(2): 2020. SJBLS.MS.ID.000509.
Shooting range, 16srRNA, Polymerase chain reaction, Bioremediation, Xenobiotic, Bacterial consortia, Microbial competition, Explosives, Enzymes, Heavy metals, Biodegradation, Prokaryotes Eukaryotes, High persistence, Toxicity, Mutagenicity
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






