 Research Article
Research Article
In vitro Characterization of SARS-CoV-2 Protein Translated from the Moderna mRNA-1273 Vaccine
Rocco Rotello, Brad Pauley, Elisha Injeti and Timothy Veenstra*
School of Pharmacy, Cedarville University, USA
Timothy Veenstra, School of Pharmacy, Cedarville University, 251 N Main StreetCedarville, OH 45314, USA.
Received Date:September 16, 2022; Published Date:September 29, 2022
Abstract
Extensive research around messenger ribonucleic acid (mRNA) vaccines and their proposed utility during the current COVID-19 pandemic resulted in many publications concerning the SARS-Cov-2 spike protein and angiotensin converting enzyme-2 receptor-binding domain of the virus, but none described what the full-length protein obtained from the modified/synthetic mRNA that is part of the Moderna and Pfizer-BioNTech vaccines. In this brief paper, we provide the first data characterizing the actual proteins produced by mouse and human cells in culture that had been incubated up to 30 minutes with the commercial vaccine produced by Moderna (i.e., Spikevax). The mRNA vaccine continues to produce proteins up to 12-14 days after introduction to the cells. The molecular weight of the SARS-CoV-2 encoded protein ranges from 135 to 200 kilodaltons depending on the extent of glycosylation.
Keywords:COVID-19; SARS-CoV-2, mRNA vaccine, in vitro expression; ELISA
Abbreviations:mRNA: messenger ribonucleic acid; FDA: Food and Drug Administration; COVID-19: coronavirus disease 19; SARSCoV- 2: severe acute respiratory syndrome coronavirus 2; ELISA: enzyme-linked immunoassay; PBS: phosphate buffered saline; REGEN-COV2: Regeneron monoclonal antibody against SARS-CoV-2; IgG: immunoglobulin; TMB: 3,3’,5,5’-tetramethylbenzidine
Introduction
The current coronavirus disease 19 (COVID-19) pandemic has resulted in several FDA approved emergency use authorizations (EUA) to slow the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,2]. Messenger ribonucleic acid (mRNA) vaccines were at the forefront of these EUAs based on their efficacy and how they could be formulated to enter cells after intramuscular injection [3-5]. RNA delivery to cells had been a challenge for decades, however, the development of lipid nanoparticles and the modification with N1-methyl-pseudouridine greatly enhanced the efficacy of both Moderna’s (Spikevax) and Pfizer-BioNTech’s (Comirnaty) mRNA vaccines [6]. In addition, modification of the mRNA with multiple N1- methyl-pseudo uridine bases enhanced the stability of the mRNA enabling it to survive the intra and extracellular environments long enough to be translated into a functional, intact protein [7]. One knowledge gap concerning these vaccines was the length of time the mRNA survives in vivo and is translated into protein. Another unknown is the size of the actual protein(s) that is translated and what does the actual protein look like after it is made within mammalian cells in culture. In this study, we evaluated the length of time and the size of the protein that the Spikevax vaccine produces in vitro. The results show that the vaccine readily enters the cells and produces a variety of protein isoforms within 24 hours. These results suggest the variability in immune response may be due to the translation of different protein isoforms that cause differences in the antibodies produced within the vaccinated or COVID-19 infected subject.
Materials and Method
Human saliva samples
Saliva samples were obtained from individuals, aged 50-65 years of age, who were (Group A) unvaccinated and contracted COVID-19; (Group B) vaccinated, had not contracted COVID-19, and tested negative for the virus; (Group C) unvaccinated, had not contracted COVID-19, and had no disease-related symptoms between September 2020 and October 2021; (Group D) vaccinated and had contracted COVID-19 based on a rapid antigen test. All samples were acquired with approval from the Cedarville University Institutional Review Board.
Saliva samples were tested from donors and volunteers at random time points using an enzyme-linked immunoassay (ELISA). Samples were collected in a collection vial containing 0.05% thimerosal. The samples were immediately centrifuged at 21,000 rpm at 4 °C to remove any particulates and stored at 4 °C. Samples being stored longer than 2 weeks contained a protease inhibitor cocktail (4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride, aprotinin, bestatin, E-64, leupeptin, and pepstatin A; Cell Signaling Technology, Danvers, MA) and were stored in aliquots at -20 °C.
Cell culture system to express SARS-CoV-2 protein(s) from synthetic mRNA-1273 vaccine (Spikevax)
Vaccine uptake was tested in the normal mouse embryonic fibroblast NIH 3T3 adherent cell line (ATCC, Manassas, VA) and human monocytic U937 cells grown in suspension. At confluence, cells were placed in DMEM or RPMI serum-free media (10 mL/100 cm dish). Two hundred microliters of room temperature Spikevax vaccine (Moderna, Cambridge, MA) was added to the cells, which were then incubated for 30 minutes with rocking at room temperature. After 30 minutes, 10% (v/v) fetal bovine serum was added, and the cells were placed in an incubator maintained at 37 °C and 4.5% CO2. Due to the nature of the lipid, cholesterol, and polyethylene glycol lipid shell that surrounds the mRNA, no other agent was required to assist the entry of the vaccine into cells. This incubation time did not affect cell viability, which remained above 90% based on trypan blue exclusion and cell counting using a CytoSMART cell counter. Cells were collected at 1, 3, 6, 12 and 24 hrs, and 5, 9, and 15 days after infection and lysed in Triton lysis buffer containing a protease inhibitor cocktail. The cell supernatants were also collected and stored at -20 °C for further analysis. Protein concentrations were measured using a Bradford microplate assay.
Enzyme-linked Immunoassays
The binding of the collected human saliva and REGEN-COV2 antibodies to cell lysates and supernatants was evaluated using an in-house developed ELISA. The reactivity of the ELISA was evaluated against the commercially available SARS-CoV-2 protein (R&D Systems, Minneapolis, MN). Briefly, Immulon TM 2 high protein binding ELISA plates (ICT; Davis, CA) were coated with SARS-CoV-2 proteins at 100 ng/well in phosphate buffered saline (PBS) at 4 °C overnight. Wells were rinsed twice with PBS and blocked using 3% blotting-grade protein blocker (Biorad). Antibodies were incubated in concentration ranges from 1-10 μg/ml and detected using donkey anti-human secondary antibodies, labeled with horse radish peroxidase (HRP) at a 1:5,000 dilution. These secondary antibodies are supplied pre-adsorbed to numerous species’ immunoglobulins (IgGs) to reduce non-specific binding. After incubation with 3,3’,5,5’-tetramethylbenzidine (TMB) substrate for 10 minutes, 50 μl of 2 N sulfuric acid was added and the absorbance of the TMB substrate at 450 nm was measured using a Promega plate reader. The ELISA was performed by coating the wells of the plate with the mRNA vaccine and probing them with the patient samples (Groups A-D) along with the commercial antibodies. No positive signals were observed in the ELISA showing that vaccinated patients do not develop auto antibodies against the vaccine.
Western blot of proteins from cells incubated with mRNA-1273 (Spikevax)
Cell lysates were quantitated and combined with 4x Laemmli buffer and heated at 100 °C for 5 minutes. The cell lysate (20 μg) was separated using a 6-18% SDS-PAGE gel. Proteins were transferred onto a nitrocellulose membrane under ice for 1 hr. Membranes were blocked using 3% blotting-grade blocker (BioRad) and washed with PBS. For Western blotting, the saliva antibodies were clarified and diluted 1:3 in blotting-grade blocker, while the REGEN-COV humanized IgG1 antibodies were dissolved at 10 μg/ml in blotting-grade blocker. Secondary antibodies were used at a dilution of 1:2,500 in 3% blotting-grade blocker. Enhanced chemiluminescence ECL was employed to detect proteins, using an Azure 600 imager.
Results and Discussion
Human saliva and REGEN-COV antibody binding to Spikevax-treated fibroblasts
Twelve saliva samples (3 from each group A-D) were analyzed in duplicate using an ELISA plate coated with the SARS-CoV-2 spike protein. Anti-SARS-CoV-2 monoclonal antibodies (REGEN COV and R&D Systems) were used as positive controls in both the ELISA and western blots. Saliva obtained from subjects in group A who presented with similar clinical profiles (i.e., unvaccinated, serious illness followed by recovery) had the strongest reactivity to the SARS-CoV-2 protein. Samples taken from Group B (vaccinated and non-COVID-19 infected) and Group D (vaccinated and previously infected with COVID-19) subjects exhibited a similar response in the ELISA, however, it was less than that observed in samples from Group A subjects. Saliva samples from Group C subjects (unvaccinated and non-COVID-19 infected) showed essentially no response, confirming the lack of anti-SARS-CoV-2 spike protein antibodies in this subject that had not been vaccinated nor infected by COVID-19. Antibodies were purified from Group A saliva samples and isotyped as strong IgG1 and IgG2 a using lateral flow strips (Thermo Fisher Scientific, Waltham, MA). While the commercial monoclonal antibodies from R&D Systems and Regeneron (REGENCOV2) showed positive reactivity in the ELISA, their response was lower than that observed using saliva samples from Group A subjects (Figure 1).
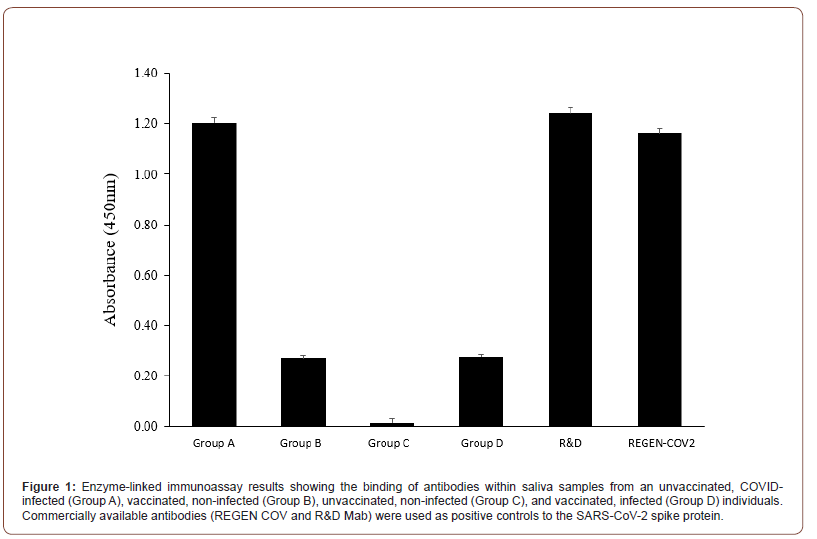
Western blots of extracts from Spikevax-treated cells
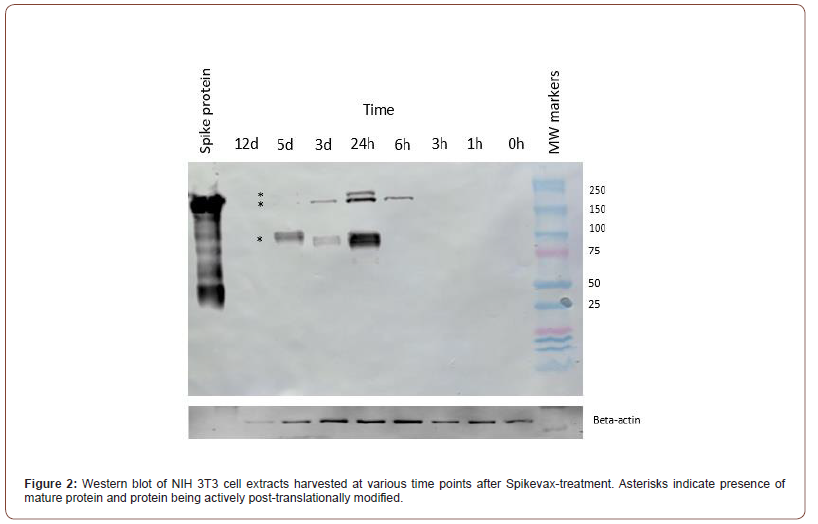
Cell lysates and supernatants were analyzed using western blotting to determine the size of protein expressed from the Spikevax vaccine mRNA. Cell lysates and supernatants collected at 1, 3, 6, 12 and 24 hrs, and 5, 9, and 15 days, were analyzed using an ELISA for presence of Spikevax-synthesized protein. Based on their strong reactivity in the ELISA study, antibodies purified from the saliva samples acquired from Group A subjects were used to perform the western blots. Western blotting of the cell lysates revealed three prominent bands at a molecular weight of approximately 180 kD (Figure 2), which can be seen most prominently 24 hours post-infection with the vaccine. The three bands with distinct molecular weights may arise from differential post-translational modifications (most likely glycosylation) that occurs as the proteins expressed from the mRNA vaccine are processed. SARS-CoV-2 protein expression was detectable in cell lysates within 6 hours of treating the cells with the vaccine (Figure 2). Protein levels peaked at 24 hours and remained detectable over 5 days. No SARS-CoV-2 spike protein was detectable in the NIH 3T3 cell lysates after 12 days. The cell supernatants did not contain any detectable vaccine-induced protein.
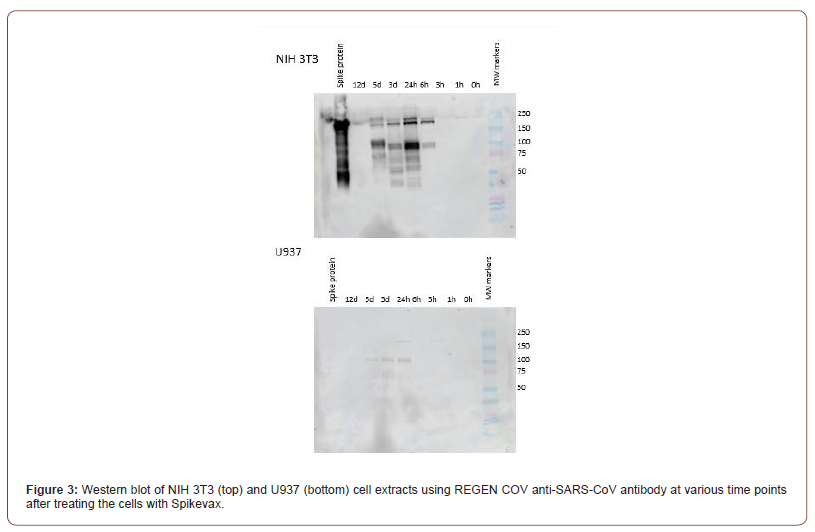
The expression of Spikevax was also analyzed in U937 cells (Figure 3). While vaccine protein expression is detectable in lysates obtained from both cell lines, spike protein levels were much lower in the U937 cells compared to NIH 3T3 cells. Detectable protein was not observed in U937 cell lysates until 24 hours after incubation with the vaccine. The numerous isoforms observed in NIH 3T3 cells were also not clearly detected in U937 cells. This reduced expression may be due to U937 cells being a leukemic cancer cell line while NIH 3T3 cells are non-tumorigenic, normal DNA and nongenetically modified [8].
Isolating mRNA is technically challenging owing to its susceptibility to degradation by many RNA- degrading enzymes and buffers [9]. This fact makes the ability to deliver an mRNA vaccine in a lipid nanoparticle vesicle through a needle into different cell types a remarkable success. The mRNA vaccine is expressed and post-translationally modified using the machinery akin to any wild-type mRNA that may be normally present within the cell. This study shows that mouse NIH 3T3 cells take up the vaccine and start expressing protein within 6 hours. The protein encoded by the mRNA vaccine is detectable within cell extracts isolated 5 days post-treatment, however, no protein is detectable within the cell supernatant at this time point. In communications with Moderna and Pfizer-BioNTech regarding the proteins expressed by their synthetic mRNA vaccines, each company’s medical information group disclosed that that they had not examined the protein dynamics more than 48 hours post-transfection in cell culture. Owing to its proprietary status, they would not disclose any information related to the nature of the protein that was expressed.
Conclusion
Both Moderna and Pfizer-BioNTech’s mRNA vaccines encode a protein with a predicted molecular weight of approximately 141 kD. The protein contains 22 predicted N-glycosylation and several O-glycosylation sites, making it heavily glycosylated [10]. Full glycosylation of these sites would probably increase the protein’s molecular weight well above 200 kD. In this study, Western blotting using a commercially available anti-SARS-CoV-2 spike protein antibody and antibodies from the saliva of non-vaccinated individuals that had been infected with COVID-19 showed three protein bands with molecular weights around 180 kD. While the exact molecular composition of these expression products is not known, the increase in apparent size compared to the predicted size,< suggests that the protein expressed from the vaccine is glycosylated. Regardless, the results show that the protein expressed from the vaccine is present in multiple isoforms within the cell. These levels of these isoforms may play a role in the differential response that individuals have to the vaccine.
Acknowledgment
The author(s) would like to thank Cedarville University for financial support during the research, authorship, and publication of this article.
Conflict of Interest
The authors declare no conflict of interest.
References
- Krause PR, Gruber MF (2020) Emergency use authorization of Covid vaccines – safety and efficacy follow- up considerations. N Engl J Med 383(19): e107.
- Quinn SC, Jamison AM, Freimuth V (2021) Communicating effectively about emergency use authorization and vaccines in the COVID-19 pandemic. Am J Public Health 111(3): 355-358.
- Andrews N, Stowe J, Kirsebom F, Toffa S, Sachdeva R, et al. (2022) Effectiveness of COVID-19 booster vaccines against covid-19 related symptoms, hospitalisation and death in England. Nat Med 28(4): 831-837.
- Patel N, Bouchard J, Oliver MB, Badowski ME, Carreno JJ, et al. (2021) Early clinical trial data and real-world assessment of COVID-19 vaccines: insights from the Society of Infectious Disease Pharmacists. Pharmacotherapy 41(10): 837-850.
- Thompson MG, Burgess JL, Naleway AL, Tyner H, Yoon SK, et al. (2021) Prevention and attenuation of Covid-19 with the BNT162b2 and mRNA-1273 vaccines. N Engl J Med 385(4): 320-329.
- Wang Y, Zhang Z, Luo J, Han X, Wei Y, et al. (2021) mRNA vaccine: a potential therapeutic strategy. Mol Cancer 20(1): 33.
- Morais P, Adachi H, Yu YT (2021) The critical contribution of Pseudouridine to mRNA COVID-19 vaccines. Front Cell Dev Biol 9: 789427.
- Denkinger DJ, Kawahara RS (1997) Expression of the vav oncogene in somatic cell hybrids. Exp Cell Res 232(2): 388-394.
- Cheng MY, Tao WB, Yuan BF, Feng YQ (2021) Methods for isolation of messenger RNA from biological samples. Anal Methods 13(3): 289-298.
- Tian Y, Parsons LM, Jankowska E, Cipollo JF (2021) Site-specific glycosylation patterns of the SARS-CoV-2 spike protein derived from recombinant protein and viral WA1 and D614G strains. Front Chem 9: 767448.
-
Rocco Rotello, Brad Pauley, Elisha Injeti and Timothy Veenstra*. In vitro Characterization of SARS-CoV-2 Protein Translated from the Moderna mRNA-1273 Vaccine. Sci J Biol & Life Sci. 2(4): 2022. SJBLS.MS.ID.000541. DOI: 10.33552/SJBLS.2022.02.000541
-
COVID-19; SARS-CoV-2, mRNA vaccine, in vitro expression; ELISA; SARS-CoV-2 Protein; Moderna mRNA; synthetic mRNA; Extracellular environments
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






