 Research Article
Research Article
Taqman Real Time PCR Assay for Desulfomicrobium Orale in Chronic Periodontal Lesions
Sebastian Becher1,5, Tilman Fritsch3,4, Marco A Vukovic4 and Wolf Dieter Grimm1,2,4*
1Witten/Herdecke University, Periodontology, Department of Dental Medicine, Faculty of Health, 58448 Witten, Germany
2Praxisteam Haßlinghausen Clinical Competence-Centre of DGParo, Germany
3Collegium Humanum, Warszawa, Poland
4NAMZahnheilkunde Bayerisch Gmain, Clinical Competence-Centre of DGParo, Germany
5MVZ Kieferchirurgie, Düsseldorf, Germany
Wolf-Dieter Grimm, Professor of Periodontology, Periodontology, Department of Dental Medicine, Faculty of Health, Witten/Herdecke University, Germany. Private Practice, Praxisteam Haßlinghausen, Clinical Competence-Centre of DGParo, Germany, Mittelstr. 70, 45 549 Sprockhövel, Germany.
Received Date: May 05, 2022; Published Date: June 03, 2022
Abstract
Introduction: Sulfate-reducing bacteria as well as Desulfomicrobium orale (D. orale) have both been shown to play a potential etiopathogenetic role for periodontal disease.
Methods: From 15 treated periodontitis patients 45 subgingival biofilm isolates (SBIs) were obtained from residual periodontal pockets ˃5mm. Real-time PCR was performed using species-specific primers and a TaqMan probe. Absolute quantification was achieved by an external recombinant DNA-standard. Sign alignment of the 16S rRNA gene was performed using commercial algorithm. Furthermore, the correlation between probing depth (PD) or clinical attachment level (CAL) and the quantity of the target gene of D. orale were analyzed.
Results: The prevalence of D. orale within the SBIs was 100%. The mean target gene amount was 7.2E+04. Sign alignment of the sequence of the 16S rRNA gene showed high similarities to further Desulfomicrobium strains. The correlation of CAL and PD with the target gene count of D. orale was not significant (p> 0.05).
Conclusion: For the first time the absolute amount of subgingival D. orale was measured using real-time PCR technology in chronic periodontal lesions. This assay provides a culture independent risk monitoring of patients suffering from periodontitis by qualitative and quantitative analysis of D. orale.
Keywords: Sulfate-reducing bacteria; Real-time PCR; Absolute quantification; Periodontal disease, Desulfomicrobium orale
Introduction
Periodontitis is a chronic inflammatory disease caused by a dysbiosis of the oral microbiota [1, 2]. The oral cavity and especially the periodontal pocket provide a unique eco-system for microbial organisms and harbors a diverse microbiota with up to 700 prokaryote species [3, 4]. Periodontal disease as a chronic infection leads to loss of the periodontium [5]. Periodontitis is initiated by subgingival bacterial biofilms whereas the destruction of the periodontium is caused by the host response [6-8]. More than 700 bacterial species can be isolated from the oral cavity, but only 50% of them can be cultured for diagnostic purposes [9]. Socransky et al. [10] established the first model for pathogenesis of periodontitis suggesting, that periodontal microbial communities can be clustered into complexes associated with disease severity. The Socransky lab identified a “red complex” harboring Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, associated with the severe form of periodontitis. Further complexes included an intermediate orange complex with, e.g., Fusobacterium nucleatum and a yellow and green complex dominated by Streptococcus species, the latter being associated with health. However, more recent concepts suggest that keystone pathogens can disrupt tissue homeostasis and change the composition of the commensal microbiota thereby generating host immune modulation and dysbiosis [11, 12]. In classic bacterial infections the diversity of the microbiota decreases as the disease develops. In most cases of periodontitis, however, the diversity of the flora increases. Conventional techniques are limited to cultivable organisms or a pre-selection of targeted pathogens is needed [10, 13]. Periodontitis has a complex polymicrobial etiology [14-17]. Currently several methods for quantification of potential periodontopathogens are used: flow cytometry, DNA-DNA Hybridization, real-time PCR [18], culturing and immunoassays [19].
Sulfate-reducing bacteria play a crucial role in natural environments. The term sulfate-reducing bacteria describes a heterogeneous group of strictly anaerobic microbes that accomplish the dissimilatory reduction of sulfate to hydrogen sulfide. This process is carried out in the absence of oxygen and therefore is called “sulfate resperation”; Sulfate replaces oxygen as an electron acceptor for energy generation [20]. In nature, sulfate-reducing bacteria play a primary role as terminal degraders of organic matters. Therefore, in periodontal lesions the bacteria depend on an environment with a highly negative redox potential, fermentation products and sulfate. By gaining little energy for cell growth SRB produce high levels of H2S. H2S is very cytotoxic for tissue and bacteria. The lethal inhalative dosis is 0.04mM [21]. In this case it is of interest that deep periodontal lesions show high concentration of H2S and Person et al. [22] could demonstrate a positive correlation of H2S and increasing pocket depth [22].
Previous studies indicated the presence of SRB in human feces, the intestinal tract, liver abscess, brain abscess and blood [23-27]. It was in 1995 that the occurrence of SRB in human periodontal pockets was reported for the first time time, van der Hoeven et al. [28]. SRB show an increased prevalence in intrabony and furcation defects [29]. Elimination of SRB in periodontal lesions after non-surgical periodontal therapy is statistically positively correlated with improved clinical parameters, whereas persistence of SRB is positively correlated with clinical signs of inflammation [30]. In 2001, Langendijk and coworkers isolated a new SRB species from periodontal lesions. Phenotypic and phylogenetic analysis indicate that these rod shaped SRB can be regarded as a new species for which they proposed the name Desulfomicrobium orale [31]. Desulfomicrobium orale sp are nonmotile, non-spore-forming Gram-negative rods 1 to 2μm in length occurring singly, in pairs, and sometimes chains. Growth occurred at 37°C with optimum pH 7 to 7.1 under strictly anaerobic conditions, with cysteine as a reducing agent. D. orale incompletely oxidizes lactate and pyruvate to acetate, using sulfate or thiosulfate as electron acceptors and releasing sulfide. In the absence of electron acceptors, pyruvate and amino acids are fermented, while lactate, peptone, mucin, and dextrose are not. Growth requires the addition of 5% Fusobacterium nucleatum cell-free spent medium (CFS).
As mentioned above the quantification of potential periodontopathogens is more fundamental in modern periodontal microbiologic diagnostic then merely qualitative analysis. Simple PCR-based assays are capable of detecting absence or presence of diseased-related bacteria but they are not quantitative [32].
The best culture independent method for the detection of microorganism is the isolation of DNA, amplification of the ribosomal DNA, cloning of the amplicon in Escherichia coli followed by sequencing of the inserted gene [33]. In numerous studies this assay was used to detect cultivable and non-cultivable spirochetes in periodontitis patients [34]. To quantify D. orale within periodontal lesions we complemented the assay above with a real-time PCR.
The purpose of the current study was to develop a culture independent TaqMan based real-time PCR to detect and quantify D. orale in chronic periodontal lesions.
Material and Methods
Subgingival biofilm isolates
45 Subgingival Biofilm Isolates (SBI) were obtained from 15 periodontitis patients taking part in a supportive periodontal treatment (SPT) program (9 male and 6 females; 58±12.3). Before the patients entered the SPT, all patients were successfully treated by scaling and root planing. All patients had been in the maintenance phase for about 6-24 month and received neither SRP nor antibiotics in the previous six months. Pocket depth (PD) and bleeding on probing (BOP) were assessed at all teeth at 6 points. Inclusion criteria were at least 3 teeth with residual PD from 5 to 8 millimeters and positive BOP. Exclusion criteria were uncontrolled systemic conditions such as diabetes or cardiac hypertension and pregnancy. From each patient 3 SBIs from 3 different teeth were obtained at sites showing PD > 5mm and positive BOP. After removal of supragingival plaque, sterile endodontic paperpoints were inserted for 20 seconds in periodontal pockets. After insertion the paperpoints were transferred into 1ml sterile DPBS (Dulbecco’s Phophate Buffered Saline) and vortexed for 30 seconds. Subsequently the paperpoints were removed and the reaction tubes were centrifugated for 1min. at 10000rpm. The supernatant was discarded, and the pellet resuspended in 200μl sterile DPBS.
DNA isolation
The isolation of the prokaryotic DNA was performed using the High Pure PCR template Preparation Kit (Roche) according to the manufacturer’s manual. The DNA was eluated in 200μl Elution Buffer (Roche).
Qualitative PCR
For further optimizing of the real-time PCR and absolute quantification via an external recombinant DNA-standard a 350bp fragment of the 16S rRNA gene was amplified using endpoint PCR. D. orale species-specific primers were used as shown in Table 1. The total volume of the reaction mixture was 50μl containing 1μl for each primer, 20μl template DNA, 0,25μl Taq Polymerase (1,25U), 5μl 10x Buffer, 1μl dntp’s and 21,7μl distilled DNAse free H20. The PCR reaction was performed using the Mastercycler-Gradient (Eppendorf, Hamburg, Germany) with the following cycling parameters: initial denaturation for 4min at 95°C followed by 35 cycles including denaturation for 30 sec. at 95°C, annaeling for 30 sec. at 57°C and extension for 30 sec. at 72°C.
Table 1:Primers for the qualitative PCR.
Post PCR analysis was performed by electrophoreses on a 1,2% agarose gel with TAE Buffer and ethidium bromide staining for visualization. A 100bp DNA-ladder (Invitrogen, Karlsruhe, Germany) was used as a molecular size standard.
Cloning of the PCR-products
The PCR-products were cloned into the Escherichia coli using the pCR®2.1-TOPO® vector (Invitrogen, Karlsruhe, Gemany) according to the manufacturer’s manual. For transformation One Shot® Competent Cells (DH5α-T1R) were used. 4μl of the ligation product were mixed with DH5α-T1R cells and gently mixed. The reaction batch was incubated for 5 minutes on ice.
After incubation 200mM NaCl and 10mM MgCl were added to optimize transformation. 50μl of the reaction batch were plated on 37°C preheated LB plates containing ampicillin and 40μl X-gal (conz. 40mg/ml) and incubated at 37°C overnight.
Bacterial culturing
Minipreperations of selected white colonies were performed in 2ml LB and 4μl Ampicillin (100mg Ampicillin/ml H20) and incubated at 37°C overnight. Isolation of plasmid DNA was performed using the Qiagen Plasmid Midi Kit according to the manufacturer’s manual (Qiagen GmbH, Hilden, Deutschland).
Restriction
Restriction of the 350bp 16S rRNA fragment was performed using 1,5μl Buffer, 10μl DNA, 0,3μl EcoR1 and 3,2μl H20 in a total volume of 15μl for one hour at 37°C. Cloning, isolation and restriction was controlled using gel electrophoresis (0,9 g agarose, 75ml TAE Buffer, 3μl ethidium bromide). A 100bp DNA-ladder (Invitrogen, Karlsruhe, Germany) was used as a molecular size standard.
Sequencing and sequence analysis
Sequencing of the PCR-product was performed according to the method established by Sanger and colleagues [35, 36] using M13 primers and the ABI-Prism 3100 Genetic Analyzer (PE Applied Biosystems, Rodgau-Jügesheim, Germany). Alignment of the 16s rRNA gene fragment of D. orale to other related microorganisms was performed using the algorithm Megablast (National Center for Biotechnology Information, Maryland, USA, http://www.ncbi.nlm. nih.gov/entrez). Alignment to Desulfovibrio fairfieldenis [26], Desulfovibrio strain NY682 (DSM 12803) [31] and Desulfovibrio desulfuricans (Devereux et al.) [37] was performed using the algorithm Blast (bl2seq) (National Center for Biotechnology Information, Maryland, USA, http://www.ncbi.nlm.nih.gov/entrez).
Midipreparations
Representative selected and sequenced clones were cultured in 50ml LB and 100μl Ampicillin and incubated at 37°C overnight. Isolation of plasmid DNA was performed using QIAfilter Plasmid Midi Kits (QIAGEN GmbH, Hilden, Germany). The supernatant was discarded, and the plasmid pellet was eluated in 50μl H2O. Adjacent, the plasmid DNA was analyzed via gel electrophoresis (E-Gel®, Invitrogen, Germany) and λHindhIII-marker (Invitrogen, Germany).
Real-time PCR for D. orale
Real-time PCR was carried out using the GeneAmp® 5700 Gene Sequence Detection System (PE Applied Biosystems, Rodgau- Jügesheim, Germany). For each real-time PCR, 9μl template DNA, 1μl each of forward and reverse primers (final concentration: 10pmol/μl), 1μl TaqMan Probe (HotStarTaq® DNA Polymerase, Qiagen, Hilden, Germany) and 12,5μl of PCR Master mix (QuantiTect PCR Master mix, Qiagen, Hilden, Germany) and 0,5μl Uracil-N-Glycosylase (1u/μl) in a total volume of 25μl was used. Each analysis was performed in triplicate. Further non-template controls (NTC) were also used. Two-step real-time PCR was performed using the following amplification conditions: UNG contamination prevention at 50°C for 2min., initial activation at 95°C for 15min., 44 PCR cycles including denaturation at 95°C for 15sec. and annealing/extension at 58°C for 60sec.
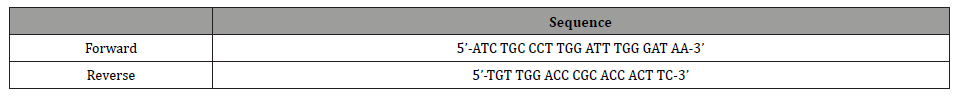
Species-specific TaqMan probe, forward and reverse primers were individually designed (Primer Express® Software v2.0, Applied Biosystems) and synthesized as shown in (Table 2) (MWG, Biotech AG, Ebersberg, Germany).
Table 2: TaqMan Probe and real-time Primers.

The critical threshold cycle (Ct) was determined in the logarithmic plot within in the log-linear phase. Baseline was set in the linear plot.
Efficiency
The real-time PCR efficiency was calculated by the formula E = 10-1/slope as described elsewhere [38].
Dilution curve
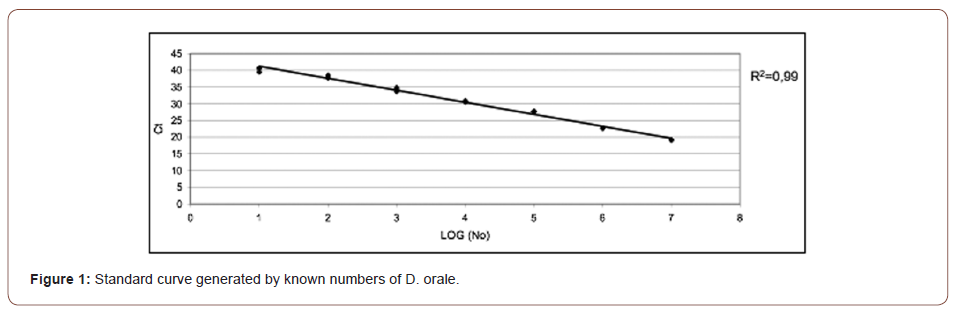
A serial dilution was generated from 101-107 gene copies/μl (see Figure 1). Hereby 16S rRNA gene copies gained from the bacterial culture were used as described above. The real-time PCR for the standard curve was performed under the identical conditions as the clinical samples. Triplicate samples were analyzed.
Absolute quantification
The absolute quantification was performed using an external recDNA standard as described previously. The generated standard curve describes the logarithm of the diluted target gene of D. orale and Ct values. The Ct value within the log linear phase is direct proportional to the logarithm of the target gene. The calculation of the target gene of D. orale was performed by means of linear regression of the standard curve and the determined slope and intercept.
Statistical analysis
Logarithmic transformation of the gene copies of D. orale was performed. These data were compared with CAL and PD by Pearson correlation analysis. Data analysis was achieved by statistic software SPSS for Windows. The significance criteria was set on p value of < 0.05.
Results
Results of sequencing and sequence analysis
The sequence analysis using the algorithm Megablast showed high similarities of the cloned and restricted fragment of the 16S rRNA gene of D. orale to other D. orale strains. The 16S rRNA gene-fragment showed a 100% similarity to D. orale strain NY676. A 99% similarity could be shown for D. orale strain NY677, D. orale strain NY 679, D. sp. oral clone BP1-74 gene, D. orale strain DSM12838 and D. orale strain NY681. For D. orale strain NY683, a 97% similarity could be demonstrated (see Table 3).
Table 3: Results of the sign alignment using the algorithm Megablast.

Results of the alignment using the algorithm Blast (bla2seq) showed a 93% similarity for D. orale to D. fairfieldensis and Desulfovibrio strain NY682 (DSM 12803). For Desulfovibrio desulfuricans 87% similarity could be demonstrated (see Table 4).
Table 4: Results of the sign alignment using the algorithm Blast (bl2seq).

Standard curve
The standard curve is shown in (Figure 1). The calculated efficiency was 1.9. The coefficient of determination was 0,99 concerning the regression line. The detection limit of the real-time PCR was 101 gene copies within the reaction tube.
Absolute quantification
A prevalence of 100% could be demonstrated for D. orale in all 45 biofilm samples.
For absolute quantification mean values of the triplicates were calculated.
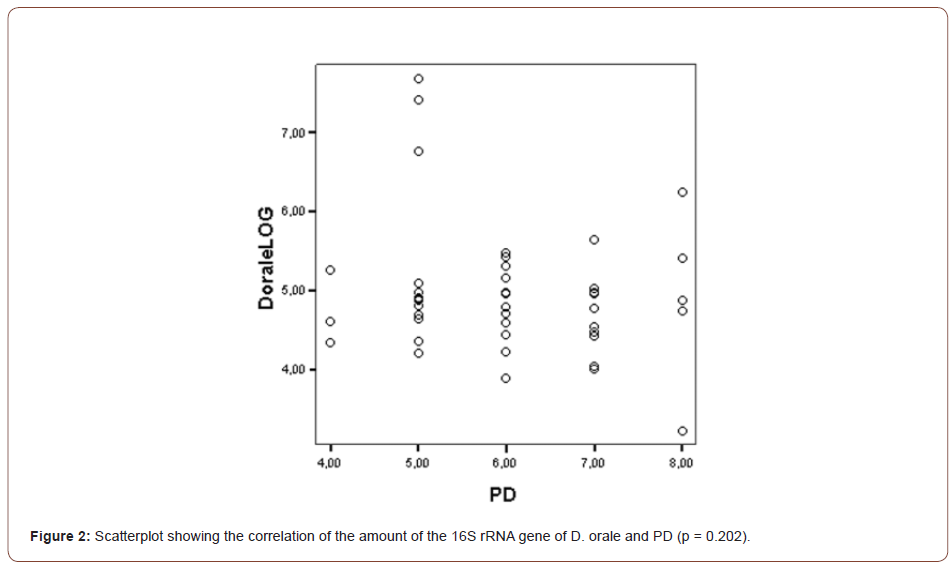
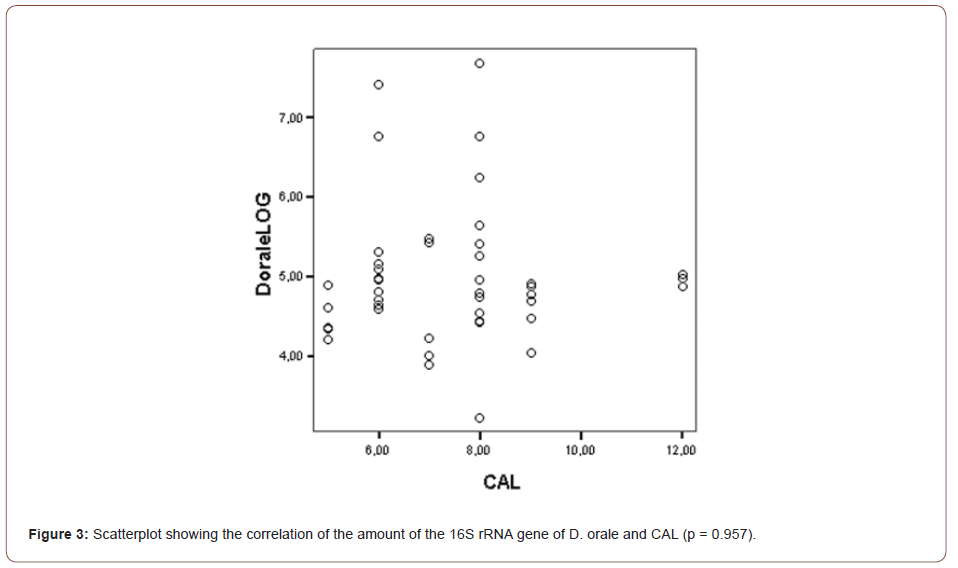
The mean gene copies of D. orale over all SBIs was 7.2E+04. The correlation of PD and CAL with the gene copies of D. orale was found not to be statistically significant (PD: p = 0.202, CAL: p = 0.957) as can be seen in (Figures 2, 3).
The range of numbers of the D. orale gene copies found in the clinical samples was 7.7E+03 – 4,7E+07.
Discussion
Strict anaerobic bacteria as potential periodontopathogens most often require special techniques for detection and identification due to their stringent growth requirements [39]. Hence it is not surprising that by using standard culture techniques microbial diversity could be greatly underestimated [40]. Less than 10 species of bacteria that are associated with periodontal disease progression are yet cultivable [41]. By the development of selective and non-selective media the spectrum of culturing could be enlarged. Here we describe the isolation and characterization of previously uncultured D. orale, the first human-associated representative of its genus. As mammalian-associated microbes rarely have free-living close relatives, D. orale provides opportunities to study how bacteria adapt and evolve within a host. This sulfate-reducing deltaproteobacterium has adapted to the human oral subgingival niche by curtailing its physiological repertoire, losing some biosynthetic abilities and metabolic independence, and by dramatically reducing environmental sensing and signaling capabilities. SRB are very vulnerable to oxygen and require a redox potential below -100mV for their growth [42].
Therefore, SRB were completely overlooked in standard culture techniques [30, 42]. This problem was overcome by the development of enrichment cultures [31], however, a quantification is still limited. The sensitivity of bacterial culturing with non-selective media is about 103-104 bacterial cells, whereas PCR is an extremely sensitivite technique, able to detect even one copy of the gene of interest [43]. Advantages of real-time PCR are the high sensitivity and specificity and the possibility of a rapid and cost-effective quantitative detection of oral pathogens [41]. The additional hybridization of the species-specific TaqMan probe increases the sensitivity and specificity of TaqMan based real-time PCR assays in contrast to dsDNA Probes as SYBR Green [44, 18]. Therefore, TaqMan real- time PCR is also used to distinguish closely related periodontal species as Provotella intermedia from Provotella nigrescens and Aggregatibacter actinomycetemcomitans from Haemophilus influenzae based on the 16S rRNA gene [18]. Based on rRNA relative sequence abundance, Desulfobulbus sp. (Deltaproteobacteria) has been one of the uncultured bacteria strongly associated with advanced periodontitis [15-17]. Deltaproteobacteria have low taxonomic diversity in the human microbiome, and other species have been linked, based on relative abundance, to either oral disease (Desulfomicrobium orale and Desulfovibrio fairfieldensis) [30, 45] or intestinal and gynecologic disorders (Desulfovibrio piger and Desulfovibrio intestinalis) [46-48]. As with other oral species, it has remained unclear if those organisms play a role in the etiology of periodontitis or their increased abundance is a consequence of the disease state, with deep subgingival pockets favoring the proliferation of strict anaerobes. All free-living Desulfobulbus species are sulfate reducers, strict anaerobes, largely prototrophic, and use a variety of electron donors and acceptors. Adaptation to a host-associated lifestyle provides relative environmental stability and close interaction with other host-recruited species. These factors often lead to genome reduction, metabolic codependence/specialization, and sometimes emergence of pathogenicity through lateral gene transfer [49-51]. Previous single-cell genomic data suggested Desulfobulbus sp. is capable of sulfate reduction [52], which we used here to selectively enrich for, isolate, and characterize that organism as the first host-associated member of the Desulfobulbus genus. Its physiological, genomic, and immunomodulatory characteristics provide new insights into adaptation to the human host-associated lifestyle and the emergence of pathogenicity.
In the present study an external recombinant DNA standard was used that is used routinely for absolute quantification [53]. Analysis of the recDNA standard was performed under the identical conditions as the clinical samples. This is essential to generate valid data [54, 38]. The calculated coefficient of determination of the standard curve was 0.99 and therefore implicates a high linearity and authenticity what is in accordance with other investigations [40, 44].
Until now D. orale as a potential periodontopathogene [31] has not been quantified using molecular-biologic assays. All present studies document analysis of the prevalence of D. orale using culture techniques. The prevalence of SRB in patients taking part in a SPT program and suffering from severe chronic periodontitis measured by culture technique ranges from 33.3%-87.5% [47, 55]. In untreated periodontitis patients, the prevalence of SRB ranges from 55.5% to 84% [56, 47]. Regarding the prevalence of SRB resp. D. orale in chronic periodontal lesions, a great heterogeneity of the data becomes obvious. This may result from different microbiological methods used in different investigations. We used a highly sensitive and specific real-time PCR assay, other groups used enrichment cultures to detect SRB [28, 57, 58, 29, 30, 59]. In the present study no significant correlation of the amount of D. orale and the PD and CAL could be shown. Langendijk, et al. [55] and Van der Hoeven et al. [28] documented different results regarding the presence of SRB and PD. Van der Hoeven, et al. [28] showed no coherence of the presence of periodontal SRB and the pocket depth [28]. Langendijk, in contrast documented a statistically significant correlation between the presence of SRB and PD [55]. Recently Vianna et al. [60] published an article dealing with the quantitative analysis of three hydrogenotrophic microbial groups. Their findings document a statistically significant difference in the mean proportions of SRB in moderate and severe cases. Phylogenetic analysis of the dsrAB gene showed a 96% accordance with Desulfovibrio strain NY682 [60]. In the present study we could document an exact match (100%) with Desulfomicrobium orale strain NY676 and 99% agreements with D. orale strain NY677, D. orale strain NY679 D. sp. oral clone BP1-74 gene, D. orale strain DSM12838 and D. orale strain NY681. Therefore, in the present investigation D. orale could be detected with maximum likelihood. Sign alignment to other potential human pathogen SRB as Desulfovibrio fairfieldensis showed no match beyond 98% so that in the present investigation D. fairfieldensis could be clearly excluded (see Table 4).
Concerning the theoretical cell count of other potential periodontopathogenes present studies document heterogeneous data ranging from 2.4E+03 – 2.6E+03 for A.a. and from 9.3E+02 – 9.0E+05 for P.g. Nonnenmacher, et al. [40], Lau et al., [61] showing similarity to our findings of 7.2E+04 for D. orale.
For the first time D. orale could be absolutely quantified using a TaqMan based real-time PCR assay. The PCR assay showed a high degree of efficiency and specificity. This assay can be used for further risk assessment in periodontal progression and will lead to a better understanding of the polymicrobial periodontal biofilm.
Acknowledgement
None.
Conflict of interest
No conflict of interest.
References
- Dye BA (2012) Global periodontal disease epidemiology. Periodontol 2000 58: 10-25.
- Yost S, AE Duran Pinedo, R Teles, K Krishnan, J Frias Lopez (2015) Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Medicine 7: 27.
- Chen C, C Hemme, J Beleno, Z J Shi, D Ning, et al. (2018) Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J 12(5): 1210-1224.
- Darveau RP (2010) Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8: 481-490.
- Schatzle M, Loe H, Lang NP, Heitz-Mayfield LJ, Burgin W, et al. (2003) Clinical course of chronic periodontitis. III. Patterns, variations and risks of attachment loss. J Clin Periodontol 30: 909-918.
- Socransky SS, Haffajee AD (1992) The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol 63: 322-331.
- Offenbacher S, Heasman PA, Collins JG (1993) Modulation of host PGE2 secretion as a determinant of periodontal disease expression. J Periodontol 64: 432-444.
- Novak MJ, Donley TG (2002) Using host response modifiers in the treatment of periodontal disease. Pract Proced Aesthet Dent 14: suppl 3-10; quiz 11.
- Paster BJ, Olsen I, Aas JA, Dewhirst FE (2006) The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 42: 80-87.
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL (1998) Microbial complexes in subgingival plaque. J Clin Periodontol 25: 134-144.
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, et al. (2011) Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10: 497-506.
- Hajishengallis G, Darveau RP, Curtis MA (2012) The keystone-pathogen hypothesis. Nat Rev Microbiol 10: 717-725.
- Socransky SS, Haffajee AD, Teles R, Wennstrom JL, Lindhe J, et al. (2013) Effect of periodontal therapy on the subgingival microbiota over a 2-year monitoring period. I. Overall effect and kinetics of change. J Clin Periodontol 40(8): 771-780.
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, et al. (2013) The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7: 1016-1025.
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, et al. (2012) Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6: 1176-1185.
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, et al. (2012) Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 7: e37919.
- Oliveira RR, Fermiano D, Feres M, Figueiredo LC, Teles FR, et al. (2016) Levels of candidate periodontal pathogens in subgingival biofilm. J Dent Res 95: 711-718.
- Kuboniwa M, Amano A, Kimura KR, Sekine S, Kato S, et al. (2004) Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol Immunol 19: 168-176.
- Sanz M, Lau L, Herrera D, Morillo JM & Silva A (2004) Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: a review. J Clin Periodontol 31: 1034-1047.
- Langendijk PS (2001) Sulfate-Reducing Bacteria in Human Periodontitis. Universität Nijmegen.
- Beauchamp RO, Bus JS, Popp JA, Boreiko CJ, Andjelkovich DA (1984) A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol 13: 25-97.
- Persson S (1992) Hydrogen sulfide and methyl mercaptan in periodontal pockets. Oral Microbiol Immunol 7: 378-379.
- Gibson GR, Macfarlane GT, Cummings JH (1993) Sulphate reducing bacteria and hydrogen metabolism in the human large intestine. Gut 34: 437-439.
- Porschen RK, Chan P (1977) Anaerobic vibrio-like organisms cultured from blood: Desulfovibrio desulfuricans and Succinivibrio species. J Clin Microbiol 5: 444-447.
- Gibson GR, Macfarlane GT, Cummings JH (1988) Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J Appl Bacteriol 65: 103-111.
- Tee W, Dyall-Smith M, Woods W, Eisen D (1996) Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J Clin Microbiol 34: 1760-1764.
- Lozniewski A, Maurer P, Schuhmacher H, Carlier JP, Mory F (1999) First isolation of Desulfovibrio species as part of a polymicrobial infection from a brain abscess. Eur J Clin Microbiol Infect Dis 18: 602-603.
- Van Der Hoeven JS, Van Den Kieboom CW, Schaeken MJ (1995) Sulfate-reducing bacteria in the periodontal pocket. Oral Microbiol Immunol 10: 288-290.
- Langendijk PS, Hagemann J, Van Der Hoeven JS (1999) Sulfate-reducing bacteria in periodontal pockets and in healthy oral sites. J Clin Periodontol 26: 596-599.
- Langendijk-Genevaux PS, Hanssen JT, Van Der Hoeven JS (2001) Decrease of sulfate-reducing bacteria after initial periodontal treatment. J Dent Res 80: 1637-1642.
- Langendijk PS, Kulik EM, Sandmeier H, Meyer J, Van Der Hoeven JS (2001) Isolation of Desulfomicrobium orale sp. nov. and Desulfovibrio strain NY682, oral sulfate-reducing bacteria involved in human periodontal disease. Int J Syst Evol Microbiol 51: 1035-1044.
- Rudney JD, Chen R, Pan Y (2003) Endpoint quantitative PCR assays for Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J Periodontol Res 38: 465-470.
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA (1986) Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol 40: 337-365.
- Dewhirst FE, Tamer MA, Ericson RE, Lau CN, Levanos VA, et al. (2000) The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol Immunol 15: 196-202.
- Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94: 441-448.
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74: 5463-5467.
- Devereux R, He SH, Doyle CL, Orkland S, Stahl DA, et al. (1990) Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J Bacteriol 172: 3609-3619.
- Rasmussen RP (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K, (Eds.) Rapid Cycle Real-time PCR, Methods and Applications. Springer Press: Heidelberg, Germany.
- Yoshida A, Kawada M, Suzuki N, Nakano Y, Oho T, et al. (2004) TaqMan real-time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol Immunol 19: 196-200.
- Nonnenmacher C, Dalpke A, Mutters R, Heeg K (2004) Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods 59: 117-125.
- Boutaga K, Van Winkelhoff AJ, Vandenbroucke-Grauls CM & Savelkoul PH (2006) The additional value of real-time PCR in the quantitative detection of periodontal pathogens. J Clin Periodontol 33: 427-433.
- Postgate JR (1984) The sulphate-reducing bacteria, Cambridge University Press: Cambridge, UK.
- Greenstein G (1988) Microbiologic assessments to enhance periodontal diagnosis. J Periodontol 59: 508-515.
- Asai Y, Jinno T, Igarashi H, Ohyama Y, Ogawa T (2002) Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J Clin Microbiol 40: 3334-3340.
- Loubinoux J, Bisson Boutelliez C, Miller N, Le Faou AE (2002) Isolation of the provisionally named Desulfovibrio fairfieldensis from human periodontal pockets. Oral Microbiol Immunol 17: 321-323.
- Ichiishi S, Tanaka K, Nakao K, Izumi K, Mikamo H, Watanabe K (2010) First isolation of Desulfovibrio from the human vaginal flora. Anaerobe 16 :229-233.
- Loubinoux J, Valente FM, Pereira IA, Costa A, Grimont PA, et al. (2002) Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int J Syst Evol Microbiol 52: 1305-1308.
- Rowan F, Docherty NG, Murphy M, Murphy B, Calvin Coffey J, et al. (2010) Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum 53: 1530-1536.
- Widdel F, Pfennig N (1982) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty-acids II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch Microbiol 131: 360-365.
- Bentley SD, Parkhill J (2015) Genomic perspectives on the evolution and spread of bacterial pathogens. Proc Biol Sci 282: 20150488
- Bliven KA, Maurelli AT (2016) Evolution of bacterial pathogens within the human host. Microbiol Spectr 4: 1-8.
- Campbell AG, Campbell JH, Schwientek P, Woyke T, Sczyrba A, et al. (2013) Multiple single-cell genomes provide insight into functions of uncultured deltaproteobacteria in the human oral cavity. PLoS One 8: e59361.
- Pfaffl MW (2004) Quantification strategies in real-time RT-PCR. In: Bustin SA, (Ed.) The real-time PCR Encyclopaedia A-Z of quantitative PCR. International University Line: La Jolla, CA, pp: 87-120.
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, et al. (2001) An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25: 386-401.
- Langendijk PS, Hanssen JT, Van Der Hoeven JS (2000) Sulfate-reducing bacteria in association with human periodontitis. J Clin Periodontol 27: 943-950.
- Grimm WD, Grimm K, Hagemann J, Van Der Hoeven H, Williams RC, et al. (2003) Occurrence of sulfate-reducing bacteria (SRB) in different forms of periodontitis. German dentist Z 58: 311-315.
- Grimm WD, Arnold WH, Grimm K, Van Der Hoeven JS, Shihabi F, Langendijk-Genevaux PS (2004) Comparative in vitro studies on the adhesion of sulfate-reducing bacteria (SRB) to different resorbable barrier membranes. German dentist Z 59: 626-632.
- Willis CL, Gibson GR, Allison C, Macfarlane S, Holt JS (1995) Growth, incidence and activities of dissimilatory sulfate-reducing bacteria in the human oral cavity. FEMS Microbiol Lett 129: 267-271.
- Boopathy R, Robichaux M, LaFont D, Howell M (2002) Activity of sulfate-reducing bacteria in human periodontal pocket. Can J Microbiol 48: 1099-1103.
- Vianna ME, Holtgraewe S, Seyfarth I, Conrads G, Horz HP (2008) Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal disease. J Bacteriol 190: 3779-3785.
- Lau L, Sanz M, Herrera D, Morillo JM, Martin C, et al. (2004) Quantitative real-time polymerase chain reaction versus culture: a comparison between two methods for the detection and quantification of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in subgingival plaque samples. J Clin Periodontol 31: 1061-1069.
-
Sebastian Becher, Tilman Fritsch, Marco A Vukovic and Wolf Dieter Grimm. Taqman Real Time PCR Assay for Desulfomicrobium Orale in Chronic Periodontal Lesions. On J Dent & Oral Health. 5(5): 2022. OJDOH.MS.ID.000621. DOI: 10.33552/OJDOH.2022.05.000621.
-
Periodontal disease, Oral microbiota, Desulfomicrobium orale, Oral cavity, Periodontal pocket, periodontium, periodontopathogens, Oral subgingival, Oral disease, Chronic periodontitis
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






