 Review Article
Review Article
Temozolomide Resistance in Glioblastoma Multiforme: Mechanisms, Ramifications, and Potential Solutions to an Urgent Clinical Problem
Anthony Berdis, Department of Chemistry, Cleveland State University, 2351 Euclid Avenue, Cleveland, OH 44115, US.
Received Date: May 14, 2021; Published Date: August 25, 2021
Abstract
Glioblastoma multiforme (GBM) is the most common and malignant type of brain cancer. Current standard therapy for GBM involves surgery followed by ionizing radiation and treatment with temozolomide (TMZ), a DNA-alkylating agent. At the molecular level, TMZ produces several distinct DNA lesions such as N7 -methylguanine, N3 -methyladenine, and O6 -methylguanine that all produce cytostatic and cytotoxic effects against GBM cells. The unique molecular nature of each lesion requires that cells utilize several mutually exclusive DNA repair pathways to effectively correct the DNA damage inflicted by TMZ. At face value, the diversity in DNA repair pathways suggests that drug resistance to TMZ would be a relatively rare occurrence after treatment. Unfortunately, this is not the case. While TMZ treatment does extend post-operative survival by several months, most patients develop resistance to this anti-cancer agent which significantly reduces overall survival. This review assesses this important clinical problem by examining the molecular and cellular mechanisms responsible for the anti-cancer activity of TMZ as well as the mechanisms associated with inherent and TMZ-induced drug resistance.
Keywords: Glioblastoma multiforme; DNA damage; Chemotherapy; Drug resistance; DNA repair; Mutagenesis
Abbreviations: GBM: glioblastoma multiforme; TMZ: temozolomide; EGFR: epidermal growth factor receptor; LOH: loss of heterozygosity; PTEN: phosphatase and tensin homolog; IR: ionizing radiation; OS: overall survival; BCNU: 1,3-bis(2-chloroethyl)-1-nitrosourea; DOX: doxorubicin; MMR: mismatch DNA repair; MGMT: O6 -methylguanine methyltransferase; BER pathway: base excision repair; TLS: translesion DNA synthesis; NFκB: nuclear factor kappa B; EGFRvIII: epidermal growth factor receptor variant III; NER: nucleotide excision repair; HR: homologous recombination; DSBs: double-strand breaks; SSBs: single-strand breaks; AAG: alkylpurine-DNA-N-glycosylase; 5’-dRP: 5′ deoxyribose phosphate; PARP1: poly (ADPribose) polymerase 1; Akt-mTOR: mammalian target of rapamycin; pol η: polymerase eta; pol ɩ: polymerase iota; pol Ѳpolymerase theta; pol : polymerase zeta; pol κ: polymerase kappa; pol α: polymerase alpha; pol δ: polymerase delta; pol ε: polymerase epsilon; pol γ: polymerase gamma; pol β: polymerase beta; pol λ: polymerase lambda; pol μ: polymerase mu; TdT: terminal deoxynucleotidyl transferase; RTPCR: reverse transcriptase polymerase chain reaction; 5-NITP: 5-nitroindolyl-2’-deoxyribonucleoside triphosphate; 5-NIdR: 5-nitroindolyl-2’-deoxyribonucleoside; MTD: median time for death
Introduction
Astrocytomas arise from the glial cells (astrocytes) which serve supporting roles within the nervous system. Glioblastoma multiforme (GBM), the most common type of brain cancer, is part of the larger family of astrocytomas. As such, GBM is also classified as a grade IV astrocytoma by the World Health Organization [1,2]. GBM accounts for ~50% of total glioma cases worldwide and afflicts over 12,000 children and adults each year in the United States [3]. GBM affects males at a higher rate of incidence compared to females [4]. In addition, approximately 50% of all GBM patients are 65 years of age and older, suggesting a correlation between the occurrence of the disease with age. Finally, GBM is also the deadliest of all cancers, having a median survival timeline of ~15 months and a 10% probability of 5 years survival [5].
GBM is typically classified into two categories designated as primary or secondary. Primary GBM accounts for 90% of all GBM cases worldwide and this type confers a worse prognosis compared to secondary GBM. Primary GBM often shows molecular overexpression and amplification of epidermal growth factor receptor (EGFR), loss of heterozygosity (LOH) 10q, and mutations in p16INK4A and phosphatase and tensin homolog (PTEN) [2]. In contrast, secondary GBM contain mutations in p53 whereas such mutations are uncommon in primary GBM [2]. As described later, the lack of functional p53 adversely influences response to chemotherapeutic agents.
Standard of care for GBM currently involves surgery followed by ionizing radiation (IR) and treatment with temozolomide (TMZ). While surgery is used to reduce tumor burden, it rarely eliminates all malignant cells. Thus, it only provides a temporary solution until treatments such as ionizing radiation (IR) and chemotherapy can be applied. The current standard of treatment for initial GBM is referred to as the Stupp protocol and is named after the first author of a landmark study published in 2005 demonstrating a survival advantage for concomitant and adjuvant TMZ treatment with standard radiotherapy in patients with GBM [6]. This study involved only patients with newly diagnosed, histologically confirmed GBM and comprised a total of 573 patients with a median age of 56 years. The majority of patients (>80%) had undergone debulking surgery prior to the initiation of therapy. Patients were randomly assigned to receive radiotherapy alone (fractionated focal irradiation in daily fractions of 2 Gy given 5 days per week for 6 weeks (total of 60 Gy) or radiotherapy combined with daily TMZ (75 mg per square meter of body-surface area per day, 7 days per week from the first to the last day of radiotherapy). This was followed by six cycles of adjuvant TMZ (150 to 200 mg/m2 for 5 days during each 28-day cycle). The primary end point was overall survival (OS). After 28 months, the median OS was 14.6 months with the combination of TMZ and IR, and this was longer than 12.1 months observed with IR treatment alone. The two-year OS rate with combining TMZ with IR was 26.5%, and this was statistically higher than 10.4% obtained using IR alone. Collectively, these data showed that combining TMZ and radiotherapy resulted in a clinically meaningful and statistically significant survival benefit for newly diagnosed GBM patients without produced overt toxicities.
Unfortunately, even with aggressive treatments using IR and TMZ, the median survival time for the majority of GBM patients is less than 16 months [7]. However, several innovative treatments have recently been developed and show promise for treating GBM. For example, carmustine (1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU)) is another anti-cancer agent that alkylates and cross-links DNA during all phases of the cell cycle [8]. These modifications disrupt DNA replication and transcription to induce cell cycle arrest and/or apoptosis. Carmustine also carbamoylates proteins such as DNA repair enzymes to increase its cytotoxic effect. Since carmustine cannot effectively pass through the blood-brain barrier, it is placed on wafers (Gliadel®) and used as an intracranial implant to achieve localized chemotherapy [9,10]. Another innovative anti- cancer agent is aldoxorubicin, an analog of the anti-cancer drug doxorubicin (DOX) which is effective against many types of hematological and solid cancers [11]. DOX primarily generates double strand DNA breaks (DSBs) which are a highly cytotoxic form of DNA damage [12]. Unfortunately, it does not penetrate the blood-brain barrier and is thus not effective against GBM. However, aldodoxorubicin is a prodrug of DOX that contains a pH sensitive linker that allows for transport across the blood-brain barrier and preferential release of the drug within GBM cells [13]. In fact, results from a recent phase II clinical trial in relapsed GBM patients showed that aldoxorubicin penetrates the blood-brain barrier and is associated with objective tumor responses as judged by MRI imaging and prolonged survival [14]. Finally, the anti-angiogenic agent bevacizumab (Avastin®) has gained attention as second line therapy against GBM. However, compared to standard TMZ and radiation therapy, bevacizumab does not improve OS but rather improves the management of symptoms and overall quality of life for GBM patients [15,16].
It is beyond the scope of this manuscript to critically examine all potential therapeutic approaches used to treat GBM. As such, this review will focus on TMZ and provide readers with a fundamental understanding of the anti-cancer activities of TMZ as well as clinical problems associated with its use. Using a stepwise approach, this review will first describe the DNA lesions most commonly formed by TMZ and description of the molecular and cellular mechanisms for the anti-cancer activity of TMZ. This includes descriptions of the various repair mechanisms used to correct each DNA lesion. We will then describe how DNA repair can contribute to TMZ resistance. Finally, we will describe the ramifications of ineffectual repair focusing on the tumor recurrence and the development of drug resistance. Please note that while this review will provide the reader with some historical perspective in these areas, the focus will be on more recent articles (those published within the last 5 years) to highlight important progress in the field.
Discussion
Overview of the DNA Lesions Produced by Temozolomide
TMZ is an orally administered DNA alkylating agent that can effectively cross the blood-brain barrier [17]. This is an important feature since the blood-brain barrier represents the most significant hurdle that limits drug penetrance to effectively treat GBM and other neurological cancers. Once inside a cancer cell, TMZ produces several distinct forms of DNA damage that produce cytostatic and cytotoxic effects. A general chemical mechanism describing how TMZ causes DNA damage is provided in Figure 1. This reaction begins with the spontaneous hydrolysis of TMZ at a physiological pH of 7.4 to form a methyldiazonium ion. This product is a strong electrophile that can effectively react with nearby nucleophilic groups such as oxygens and nitrogens present on the nucleobases of DNA. In duplex DNA, the most commonly formed lesions are N7-methylguanine (70%), N3-methyladenine (10%), and O6-methylguanine (7%) [18]. While alkylation at the N1 of adenine and the N3 of cytosine can also occur, these modifications arise more frequently in single-stranded DNA rather than duplex DNA [19]. In general, all of these DNA adducts can generate anti-cancer effects.
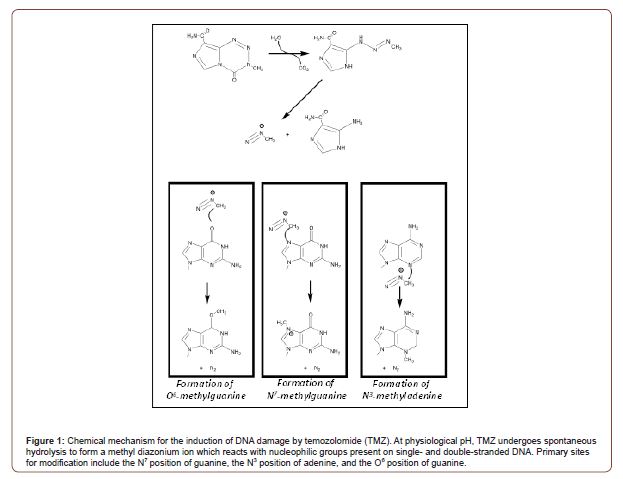
DNA damage caused by TMZ induces cellular senescence as characterized by cell-cycle arrest at the G2/M phase [20]. Aasland et al. recently showed through a series of elegant cell-based studies that TMZ-induced senescence is initiated by damage recognition through the MRN complex followed by activation of ATR/CHK1 kinases and mediated by degradation of CDC25c [21]. However, TMZ-induced senescence is also dependent upon functional p53 in addition to sustained induction of p21. While the NFκB pathway is also required for TMZ-induced senescence, neither p14 nor p16 (which are targets of p53) do not appear to play essential roles. In addition, TMZ treatment represses expression of several mismatch DNA repair (MMR) pathway proteins including MSH2, MSH6, and EXO1. Surprisingly, the homologous recombination protein RAD51 is also downregulated. Both effects appear to be dependent upon p53 activity since their repression is not observed in p53-deficient cells.
At the molecular level, the structure of each major DNA lesion produced by TMZ is different. As a result, the mechanisms accounting for their cytotoxic effects also differ. Likewise, there are multiple yet distinct DNA repair pathways that can efficiently correct DNA lesions produced by TMZ. The primary repair pathways include direct repair of O6-methylguanine by O6-methylguanine methyltransferase (MGMT), the MMR pathway, and the base excision repair (BER pathway). DNA repair can be viewed as a double edge sword. On one hand, these pathways serve to protect the genome of normal cells by correcting DNA lesions that arise from endogenous and exogenous sources. On the other hand, their activity in cancer cells can have deleterious effects on patient responses by promoting resistance to drugs such as TMZ [22-24]. In these cases, overexpression of key repair proteins is a common mechanism. In addition, loss of function via mutagenesis and/or deletion can likewise contribute to oncogenesis and drug resistance. In particular, defects in the MMR or BER pathways can allow DNA lesions caused by TMZ to persist which then provides opportunities for their mis-replication by various DNA polymerase. This process, known as trans lesion DNA synthesis (TLS), can drive drug resistance and/or mutagenesis [25-27]. To provide better insight into these activities, the sections below provide detailed descriptions of the most commonly formed DNA lesions and an overview of the DNA repair pathways used to correct these lesions. Specific attention is paid to how these repair pathways participate in generating TMZ-resistance in addition to new approaches being used to combat this clinical problem.
Cellular Effects of O6-Methylguanine
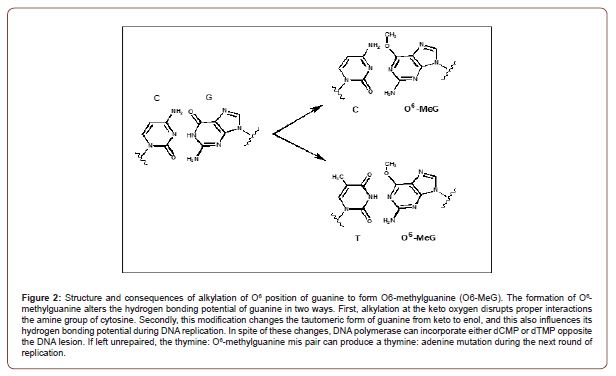
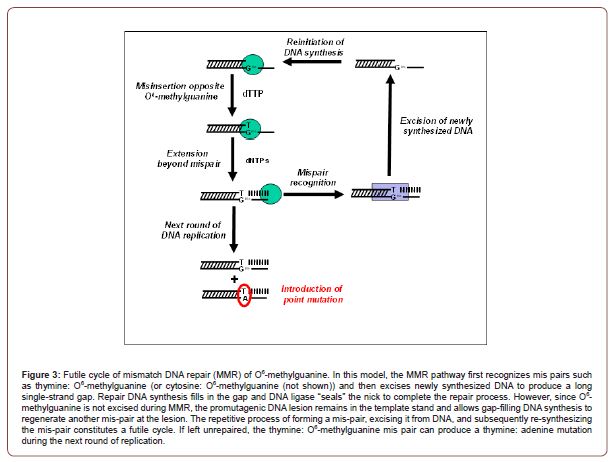
The cytotoxic effects of a methylated nucleobase are typically caused by the ability of the formed adduct to transiently disrupt the continuity of DNA replication or to block this process completely [28]. Methylation of guanine at the O6 position produces O6-methylguanine and represents an excellent example of how a DNA lesion not only disrupts the continuity of DNA synthesis but also activates DNA repair to ultimately cause cell death. As illustrated in Figure 2, methylation at the O6 position of guanine alters its hydrogen bonding potential in two ways. First, alkylation at the keto oxygen disrupts proper interactions the amine group of cytosine. Secondly, this modification changes the tautomeric form of guanine from keto to enol, and this also influences its hydrogen bonding potential during DNA replication. In spite of these changes, O6-methylguanine does not block DNA synthesis but rather allows a DNA polymerase to incorporate either cytosine or thymine opposite the lesion. Since neither nucleotide correctly base-pairs with O6-methylguanine, each mis-pair is processed by the mismatch repair (MMR) pathway [29]. In brief, the MMR pathway functions by first excising the newly synthesized DNA to produce a long single-strand gap. Repair DNA synthesis then fills in the gap and DNA ligase “seals” the nick to complete the repair process. However, O6-methylguanine it is not excised during MMR and thus remains in the template stand. As a result, gap filling DNA synthesis simply generates another mis-pair at the lesion thus creating a futile cycle that encompasses formation of a mis-pair, its excision from DNA, and subsequent re-synthesis of the mis-pair (Figure 3). Eventually, a single-strand gap forms as a result of this repetitive futile cycle which produces a DSB during the next S-phase of the cell cycle to induce cell death [30].
Repair Pathways for O6-Methylguanine: While there are several pathways that can convert O6-methylguanine back to guanine, the most studied pathway is catalyzed by MGMT, an enzyme that directly corrects O6-methylguanine by transferring the methyl group from the O6 position of guanine to an active site cysteine residue [31]. This reaction inactivates MGMT and limits it to a single-round of DNA repair rather than catalyzing multiple rounds of nucleobase correction [32]. Despite this limitation, however, MGMT activity is important for maintaining genomic fidelity as well as preventing possible tumorigenesis caused by the pro-mutagenic replication of the alkylated nucleobase.
While the importance of MGMT activity in a normal cell is intuitively obvious, it also plays a critical role in generating TMZ-resistance in GBM patients. For example, roughly 30% of all gliomas lack MGMT and thus display variable degrees of resistance to DNA damaging agents [33,34]. Expression of the MGMT gene, located in chromosomal position 10q26, is heavily modulated by several transcription factors, including nuclear factor kappa B (NFκB) and specificity protein 1 [35]. However, several studies have shown that loss of MGMT expression is not due to gene deletion, rearrangement, mutation, or unstable RNA but rather by methylation of CpG islands within the MGMT promoter [36,37]. One of the first reports of this phenomenon came from the laboratories of Baylin and Herman who collectively showed that the MGMT promoter was methylated in 19 of 47 patients (40%) of glioma patients examined [38].
Today, the methylation status of the MGMT promoter is widely used as a biomarker to predict the prognosis of GBM patients [39- 41]. In general, MGMT promoter methylation is a more accurate prognostic factor compared to other patient features such as age and tumor grade. Surprisingly, methylation of the MGMT promoter is associated with tumor regression in addition to prolonged overall and disease-free survival in response to chemotherapy. For example, Hegi et al. showed that MGMT promoter methylation correlates with better outcome in patients receiving TMZ [42]. In their study, the median OS in patients with MGMT promoter methylation was 18.2 months and significantly longer than patients without MGMT promoter methylation (median OS of 12.2 months). Likewise, Morandi et al. reported significant differences in OS using a combination of TMZ and IR [43]. This study showed a nearly 2-fold increase in median OS in GBM patient with methylated MGMT pro
moters (36 months) compared to GBM patients with unmethylated MGMT promoters (20 months). This conclusion is supported by a recent report by Kim et al. showing similar survival results in GBM patients [44]. Overall, these and other studies show that methylation of the MGMT promoter in GBM may be a useful predictor of the responsiveness of the tumors to TMZ treatment.
It should be noted that despite strong evidence correlating MGMT methylation status with survival of GBM patients, there are challenges in routinely applying this information in clinical settings. As described in a review by Mansouri et al., there remain significant controversies involving proper methods and optimal cutoff definitions for determining MGMT methylation status [45]. In particular, variations in detection methods between laboratories are a major challenge. Furthermore, Thomas et al. have argued that examining DNA methylation of the MGMT promoter may not directly or accurately reflect MGMT expression and/or activity [46]. As discussed below, there is an inverse correlation between the expression of certain MMR proteins in relation to MGMT expression. As such, it is argued that MGMT activity and MMR status should be examined concomitantly to better define prognostic indications for responses to TMZ. Finally, consideration of other clinical and genetic/epigenetic factors may also be important for optimal treatment decisions.
In addition to providing important prognostic information, the clinical benefit for the loss-of-function in MGMT activity has important ramifications in effective treatment strategies against GBM. In particular, molecules such as O6-benzylguanine have been used to inhibit MGMT activity which then sensitizes cancer cells to the cytotoxic effects of DNA alkylating agents [47-51]. Friedman at el. were amongst the first to examine if resistance to DNA alkylating agents in drug-resistant human GBM (D-456 MG) can be reversed by coadministration of O6-substituted guanines such as O6-methylguanine and O6-benzylguanine [51]. This was examined in xenografts grown subcutaneously in athymic BALB/c mice. Response to these combinations were assessed by quantifying tumor growth delay and tumor regression in addition to measuring MGMT activity at different times post-treatment. GBM xenografts were completely resistant to BCNU when treated with a dose of BCNU equivalent to its LD10 value. However, pretreatment with O6-benzylguanine increased BCNU sensitivity whereas treatment with BCNU and O6-methylguanine had no effect on either tumor regression or growth. These anti-tumor effects mirrored the ability of each compound to inhibit MGMT activity. In particular, pretreatment with O6-benzylguanine had a more pronounced inhibitory effect against MGMT activity (~90% inhibition at 6 hours post-treatment) compared to pretreatment with O6-methylguanine (~20% inhibition at 6 hours post-treatment). Subsequent studies have recapitulated these results when combining O6-benyzlguanine with TMZ. For example, Wedge and Newlands demonstrated that xenografts treated with 40 mg/kg of O6-benyzlguanine alone did not show reductions in tumor growth whereas pretreatment with O6-benyzlguanine significantly enhanced the anti-tumor activity of TMZ [52]. These authors also confirmed that pre-treatment with O6-benyzlguanine significantly inhibited MGMT after 24 hours post-treatment. Finally, a phase II trial was performed to examine if inhibiting MGMT activity by using O6-benzylguanine could improve the overall efficacy of Gliadel wafers [53]. In this study, the primary end point for 6-month OS was 82% while secondary endpoints examining 1-year and 2-year, OS were 47% and 10%, respectively. These data suggest that efficacy of implanted Gliadel wafers may be improved with coadministration of O6-benzylguanine. However, there are several safety concerns as systemic administration of O6-benzylguanine with Gliadel wafers may increase the risk of hydrocephalus, CSF leak, and CSF/brain infections.
Mismatch DNA Repair
Chemoresistance in GBM has been largely attributed to repair of TMZ-induced DNA lesions by MGMT activity. However, some MGMT- deficient glioblastomas are still resistant to temozolomide, thus indicating that other mechanisms must exist to account for drug resistance. As noted earlier, the mismatch repair (MMR) pathway also plays an important role in modulating cellular response to O6-methylguanine. The MMR pathway primarily corrects replication errors that form during chromosomal replication [54]. Mismatches are classified as base–based mis pairs (i.e., T paired opposite G) or insertion / deletion mismatches that form as a result of strand slippage at repetitive DNA sequences. In humans, the MMR pathway features two families of MMR proteins that are heterodimeric homologs of bacterial MutS (MSH) or MutL (MLH). Both homologs have been extensively examined through biochemical and cell-based studies [55-57]. The MMR pathway is divided into three distinct phases - mismatch recognition, excision, and re-synthesis of excised DNA. Each phase is catalyzed by a confederation of proteins, and their activities are tightly regulated to ensure complete and accurate repair of damaged DNA.
The first stage of MMR is mismatch recognition by MutSα or MutSβ, and the choice of protein depends upon the type of DNA damage. For example, MutSα initiates repair of base–base mispairs and small deletions of 1 to 2 nucleotides while MutSβ initiates repair of DNA containing larger deletions (1 to 15 nucleotides). In either case, binding of MutSα or MutSβ to a mismatch induces an exchange of ADP for ATP, and this exchange is necessary to convert either protein into a “sliding clamp” that scans DNA to interact with MutLα and possibly identify other mismatches.
The second phase, DNA excision, is the process in which newly synthesized DNA strand that contains the error (C or T opposite O6-methylguanine) is excised while leaving the templating strand unaltered. In prokaryotes, strand recognition is dictated by the unmethylated strand of hemi methylated DNA that occurs for a brief time after DNA replication before it is converted into fully methylated DNA. While strand recognition is not completely understood in eukaryotes, recent results suggest that PCNA plays an important role in regulating this process [58]. In vitro and in vivo evidence demonstrate that PCNA can interact with MutS and MutL in addition to other proteins such as EXO1, Pol δ, and Pol ε that are involved in re-synthesizing excised DNA [59-61]. In vitro MMR reactions require a preexisting nick or single-strand gap in the heteroduplex DNA substrate that can reside on the 5′- or 3′- side of the mismatch. In 5′-MMR reactions, excision is catalyzed by EXO1, a 5′ to 3′ exonuclease that is activated by MutSα in a mismatch-dependent manner. Excision in the case of 3′-MMR (a break or gap located on the 3′ side of the mismatch) is less well understood since EXO1 is currently the only exonuclease that appears to be involved in MMR.
The third and final phase of MMR involves DNA gap filling by a high-fidelity polymerase followed by ligation catalyzed by DNA ligase I. These activities collectively generate a corrected and intact DNA duplex. During this process, PCNA is loaded at a 3′ terminus by RFC, and replicative DNA polymerases, Pol δ, and Pol ε, replicate the RPA-coated gapped DNA. DNA ligase I generates the final phosphodiester bond to fully repair the DNA.
MMR has been implicated in TMZ resistance through the development of MMR mutations both de novo and in response to standard chemotherapeutic treatment. These MMR mutations have been found to lead to hypermutation in recurrent tumors, particularly in the setting of initial MGMT methylation [62]. Surprisingly, there appears to be an inverse correlation between the expression of MMR proteins in relation to MGMT expression. with the combination of methylated MGMT and high MMR activity conferring the best response to TMZ [63]. This relationship may be exploited in the development of TMZ resistance, where a protective DNA repair genotype may be counteracted by a mutation in a different repair system.
MMR genes including MLH1, MSH2, and MSH6 may play pivotal roles in tumor response to TMZ treatment. Friedman et al. evaluated TMZ responses in patients with newly diagnosed malignant glioma as well as the predictive value of quantifying tumor MMR and MGMT activity in these patients [64]. Thirty-three patients with newly diagnosed glioblastoma multiforme (GBM) were treated with TMZ at a starting dose of 200 mg/m2 daily for 5 consecutive days with repeat dosing every 28 days after the first daily dose. Immunochemistry was used to detect DNA mismatch repair proteins, MSH2 and MLH1, using monoclonal antibodies. Tumor samples were characterized with respect to percent positive staining. Of the 33 patients with GBM, complete responses were observed in only in three patients (9%), partial responses occurred in 14 patients (42%), stable disease was achieved in four patients (12%), and 12 patients (36%) developed more progressive disease. Thirty tumors isolated from these patients were examined for positivity for MSH2 and MLH1. Surprisingly, there was no direct correlation between levels of either MMR protein with patient responses to TMZ. Likewise, a recent study by Stark et al. using immunohistochemical staining on paraffin-embedded specimens from initial tumors of over 200 GBM patients showed that expression of MLH1, MSH2, and MSH6 in initial GBM is not associated with patient survival [65].
While MMR activity may not play important roles in mediating responses to TMZ in initial GBM, there is some evidence that MMR status may be significant in treating recurrent GBM. For example, Indraccolo et al. examined the expression of MMR proteins using immunohistochemical staining in GBM samples collected from adult patients at the time of their diagnosis and at the time recurrence after treatment with the Stupp protocol [66]. A key feature of this study was that whole exome sequencing was performed on tumor samples showing loss of MSH6 reactivity. Their results showed that 96% of GBM samples at diagnosis expressed MMR protein. However, expression of MMR proteins was partially or completely lost in ~25% of recurrent GBM samples. In addition, MSH6-negative samples had high tumor mutational burden as well as significant telomere shortening and MGMT methylation.
This least feature is highlighted by a recent study by Struve et al. examining the role of the epidermal growth factor receptor variant III (EGFRvIII), an oncogene frequently expressed in GBM, toward driving drug resistance [67]. This group performed a retrospective analysis examining OS of more than 300 GBM patients treated under the Stupp protocol. Their analysis showed that EGFRvIII expression correlates with increased OS, but that this correlation is only observed in GBM patients with MGMT promoter methylation. Molecular analyses examining EGFRvIII expression in isogenic GBM cell lines showed that EGFRvIII increases cellular sensitivity to TMZ. This favorable phenotype reflects increased DSB formation which subsequently induces cell cycle arrest at the S/G2- phase. Furthermore, both EGFRvIII-positive cells and patient tumor samples showed higher expression levels of MSH2 and MSH6. Collectively, these results suggest that the EGFRvIII sensitizes GBM cells that are MGMT deficient by upregulating the expression of key proteins involved in the MMR pathway.
Based upon these complexities, there have been several studies aimed at developing a more comprehensive assessment of DNA repair capacity that encompasses MGMT, MMR, BER, nucleotide excision repair (NER), and homologous recombination (HR) as mechanisms for drug resistance. The translational goal is to quantify global DNA repair capacity in tumor samples in order to identify patients that are most likely to respond to specific DNA damaging agents. The group led by Samson recently developed a fluorescence-based multiplexed host cell reactivation assay that can measure DNA repair catalyzed by multiple pathways [68]. Using this assay in combination with drug sensitivity measurements, this group examined 12 patient-derived xenograft models of GBM to predict glioblastoma response to TMZ treatment. There results showed changes in MGMT activity as well as small changes in MMR, NER, and HR capacity that contribute to acquired TMZ resistance in these models. Thus, quantifying the collective activity of MMR, HR, NER, and MGMT may provide a more accurate prognostic indicator of TMZ resistance compared to individual assessments of MGMT activity.
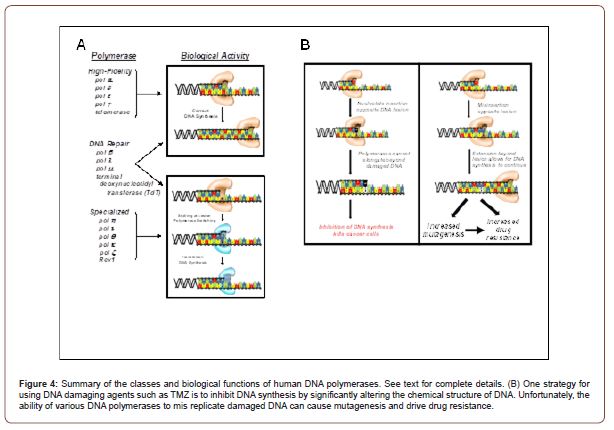
Cellular Effects of N3-Methyladenine and N7-Methylguanine: The alkylated DNA lesions, N3-methyladenine and N7-methylguanine are commonly formed by TMZ treatment and represent unique challenges to cellular proliferation and viability. In contrast to methylation at the O6 position of guanine which does not inhibit DNA replication, methylation at the N3 position of adenine directly inhibits DNA synthesis catalyzed by several DNA polymerases. This occurs since this modification disrupts essential contacts between adenine and critical active site amino acid residues with active site of the DNA polymerase to block DNA synthesis (Figure 4) [69-71]. This inhibition causes the formation of blocked replication forks that are unstable and ultimately collapse to produce double-strand breaks (DSBs) which are highly cytotoxic.
N7-methylguanine would appear to be a rather harmless modification as it occurs at a position that does not directly influence DNA polymerase activity. However, methylation at this position (as well as the N3 position of guanine and the N7 position of adenine) accelerates the rate of non-enzymatic hydrolysis of the glycosylic bond between the nucleobase and deoxyribose [72,73]. This depurination reaction forms a new distinct lesion termed an abasic site which functions as a strong block to DNA synthesis catalyzed by most DNA polymerases [74-76].
Base Excision Repair Pathway
The most abundant TMZ-induced adducts, N3-methyladenine, N7-methylguanine, and a basic site are processed by the short patch base excision repair (BER) pathway. The BER pathway primarily functions to repair endogenous forms of DNA damage caused by reactive oxygen species generated during oxidative metabolism [77]. Spontaneously generated forms of damaged DNAs include oxidized bases (8-oxoguanine), abasic sites, and single-strand breaks (SSBs). In a normal cell, it is estimated that as many as 50,000 BER-repaired lesions form per cell during the course of a single day [78]. However, treatment with TMZ increases the number of these lesions by several orders of magnitude and puts a remarkable strain on the efficacy of BER.
The repair of N3-methyladenine and N7-methylguanine is initiated by recognition of the lesions by alkylpurine-DNA-N-glycosylase (AAG). AGG cleaves the glycosylic bond between the damaged base and deoxyribose to produce an abasic site as a key intermediate in the repair process. After the hydrolysis reaction, AAG remains bound to the abasic site and recruits the endonuclease, Ape1, to the site of damage. Ape1 cleaves the DNA phosphodiester backbone to produce SSB containing a 3′-OH and a 5′ deoxyribose phosphate (5’-dRP) termini. Ape1 is then replaced by DNA polymerase β, a repair DNA polymerase that possesses 5′ lyase activity that can excise the 5′-dRP to produce a single nucleotide gap. The formed gap is subsequently filled in by DNA polymerase β, and DNA ligase seals the nick to finalize the repair process.
Several proteins involved in the BER pathway including DNA glycosylase MPG and DNA polymerase-β (pol-B) are associated with promoting resistance to TMZ and confer poor prognosis in patients [79,80]. Another protein involved in the BER pathway is poly (ADP-ribose) polymerase 1 (PARP1) which recognizes breaks in single stranded DNA and protects cells from accumulating too many apurinic/apyrimidinic sites [80,81]. Inhibitors of PARP1 have been shown to increase the cytotoxicity of TMZ in several cancer types including gliomas. For example, Higuchi et al. tested whether PARP1 inhibition could re-sensitize MSH6-null MMR-deficient gliomas to TMZ [82]. In their studies, treatment with the PARP1 inhibitor, veliparib, had no detectable effect against wild-type GBMs. However, treatment with veliparib restored TMZ sensitivity in MSH6-deficient GBM cells. In addition, combining veliparib and TMZ suppressed tumor growth in MSH6-inactivated xenografts X-fold more effectively compared to treatment with TMZ alone. Another study by Tang et al. [83] found that elimination of polcombined with the administration of PARP1 inhibitors increases TMZ sensitivity. The cause in this case appears to be a build-up of damaged DNA. Collectively, these findings suggest that mutations in components that either regulate or participate in the BER pathway can increase the potential of TMZ cytotoxicity. Finally, it is interesting to note that PARP1 activity can play an alternative role in mediating drug resistance as overstimulation of PARP1 can actually promote more DNA damage as well as cause a depletion in both NAD+ and ATP [84-86].
Translesion DNA Synthesis as a Resistance Mechanism
Common mechanisms associated with TMZ resistance involve alterations in DNA repair pathways including as MGMT and MMR. The lack of efficient DNA repair makes cancer cells more susceptible to mutagenesis which can in turn accelerate the development of additional mechanisms that drive drug-resistance. For example, a study by Johnson et al. showed that genomic DNA isolated from TMZ-treated tumors contained ~30 to 90 mutations per mega base compared to initial, untreated tumors which had significant lower mutation frequencies of <4 mutations per mega base [87]. These recurrent GBM tumors were also drug resistant due to the accumulation of mutations in genes associated with MMR, retinoblastoma, and mammalian target of rapamycin (Akt-mTOR) pathways [87]. Collectively, these results highlight the negative consequences in which unrepaired DNA lesions are mis replicated by various DNA polymerases in a biological process known as trans lesion DNA synthesis (TLS).
TLS in humans has been historically attributed to the activity of a family of DNA polymerases referred to as “specialized” due to their ability to replicate various DNA lesions with high efficiency and fidelity (Figure 4). Humans possess at least six different specialized polymerase that include polymerase eta (pol η), polymerase iota (pol ι), polymerase theta (pol Ѳ), polymerase zeta (pol ζ), polymerase kappa (pol κ), and the Rev1 protein. This unique activity directly contrasts that of “high-fidelity” DNA polymerases such as polymerase alpha (pol α), polymerase delta (pol δ), polymerase epsilon (pol ε), telomerase, and polymerase gamma (pol γ) which are involved in chromosomal and mitochondrial DNA synthesis. These polymerases display optimal activity on undamaged DNA and low activity with damaged DNA. However, there are reported instances in which high-fidelity polymerases can efficiently replicate small, miscoding DNA lesions such as O6-methylguanine and 8-oxo-guanine [88-90]. Another class of DNA polymerase are those involved in DNA repair pathways and include polymerase beta (pol β) which is responsible for processing damaged DNA repaired during base excision repair, polymerase lambda (pol λ) and polymerase mu (pol μ) which repair DSBs formed during homologous recombination and non-homologous end-joining, and terminal deoxynucleotidyl transferase (TdT) that randomly incorporates nucleotides into single strand DNA during V(D)J recombination [91,92].
Aberrant TLS activity, particularly catalyzed by pol ɩ, and pol η, is associated with tumorigenesis and drug-resistance in GBM [93- 95]. For instance, Wang et al. quantified expression levels of pol κ, pol ɩ, and pol η by quantitative RT PCR and Western blot analysis in 40 primary glioma samples and 10 normal brain samples [93]. Glioma samples showed no significant expression of pol η while overexpression of pol ɩ and pol κ was frequently observed in patient samples (~28% and ~58%, respectively). The prognostic value of these results was confirmed using a population-based tissue microarray derived from a cohort of 104 glioma patients. Immunohistochemical studies showed positive pol κ staining in ~32% of specimens while ~70% were positive for pol κ staining. More importantly, an inverse correlation between increased pol κ- and pol ι-positive staining with reduced survival was observed within this set of glioma patients. The same group also showed that overexpression of pol κ in TMZ-sensitive GBM cells conferred resistance to the drug while lowering pol k expression sensitized cancer cells to TMZ. In this case, pol κ depletion caused a cascade of cellular events such as defects in homologous recombination-mediated repair and restart of stalled replication forks.
It should be noted that two high-fidelity DNA polymerases, pol ε and pol δ, may also play significant roles in the initiation and progression of brain cancers [79]. For example, a recent study by from Campbell et al. assessed mutation burden in >81,000 tumors from pediatric and adult patients through DNA sequencing analysis [96]. Their findings show that a large number of pediatric cancers possess defects in mismatch repair pathway genes POLE and POLD1. In addition, pediatric tumors such as leukemia/lymphomas and malignant gliomas possessed greater than 100 mutations per meg abase (ultra-hypermutated) and were all replication repair deficient. In these cases, the authors conclude that while defects in MMR alone can initiate tumorigenesis by increasing mutagenesis, it is the combination of defective MMR coupled with mutations in pol δ and pol ε that cause the hyper-mutated state observed in these cancers.
Approaches to Combat Trans lesion DNA Synthesis
Cancers such as GBM typically possess defects in one or more DNA repair pathways. These defects give a cancer cell only one option to survive the effects of TMZ – it must replicate damaged DNA inflicted by this agent. Thus, one hypothesis is that inhibiting the ability of DNA polymerases to mis replicate DNA lesions that have escaped DNA repair could improve patient outcomes. On recent report showed that an artificial nucleotide analog, designated 5-NITP, is efficiently incorporated opposite a basic site, the most toxic DNA lesion generated by TMZ [97]. Results from in vitro assays showed that high-fidelity DNA polymerases such as pol δ and pol ε insert 5-NITP opposite the lesion ~100-times more efficiently than the preferred natural substrate, dATP [97]. Specialized DNA polymerases including pol η and pol ɩ also insert 5-NITP opposite an abasic site with 100-fold higher efficiencies compared to dATP. While 5-NITP is efficiently inserted opposite an abasic site, it is refractory to elongation and thus acts as chain terminator against TLS activity.
Cell-based studies using U87 cells as a human model for GBM showed that combining the corresponding nucleoside analog, 5-NIdR, with TMZ significantly increases the cell killing effects of the DNA damaging agent. as reflected in 4-fold higher levels of early and late stage apoptosis compared to cells treated individually with TMZ or 5-NIdR [97]. Flow cytometry experiments showed that treating U87 cells with 100 M TMZ induced cell-cycle arrest at G2/M with a concomitant reduction in cells at G0/G1. These changes occur with minimal effects on cells at S-phase or through increases in sub-G1 DNA levels. However, combining 5-NIdR with TMZ caused a block at S-phase. Since treatment with 5-NIdR alone has no effect on cell cycle progression, this block at S-phase likely represents the ability of the artificial nucleoside to inhibit TLS activity during S-phase. Furthermore, the increase in sub-G1 DNA observed with this combination suggests that cell accumulating at S-phase undergo cell death via mitotic catastrophe. Finally, pre-clinical testing in a heterotopic xenograft mouse model showed that 5-NIdR effectively increases the therapeutic efficacy of TMZ. These studies showed that the median time for death (MTD) for mice treated with TMZ alone was 45 days whereas the MTD for mice treated with TMZ and 5-NIdR was greater than 250 days. Equally important, >70% of mice treated with this combination showed complete tumor regression with 30 days post-treatment. Finally, initial toxicology studies show that repeat dosing with 500 mg/kg produces no adverse hematological effects or any effects of organs including brain, heart, liver, and kidney.
Conclusion
Drug resistance to DNA damaging agents such as TMZ is a significant problem in the treatment of all cancers. However, it is arguably the most significant issue toward effectively treating patients with GBM. TMZ is the most widely used anti-cancer agent against GBM. When combined with radiotherapy, TMZ provides a significant survival benefit for newly diagnosed GBM patients. However, TMZ-resistance occurs very rapidly, and this produces a poor prognosis for GBM patients due to the limited number of anti-cancer agents that can effectively cross the blood brain barrier. The most common mechanisms associated with TMZ-resistance involve alterations in DNA repair pathways such as MGMT and MMR. In some cases, increased MGMT activity drives resistance by repairing the damage inflicted by TMZ while decreased MGMT levels via epigenetic mechanisms or through the use of pharmacological inhibitors can reverse TMZ-resistance. Likewise, a lack of MGMT activity coupled with loss of MMR activity also generates TMZ resistance. These combined defects allow lesions generated by the TMZ to persist and subsequently replicated by different DNA polymerases. Since this activity can be highly pro-mutagenic, it can increase mutational frequencies to produce more aggressive cancers and/or cause tumor recurrence. A new approach to combat this problem is to block the ability of DNA polymerases to replicate lesions produced by TMZ.
Acknowledgement
This work was supported by private donations to the Berdis Glioblastoma Fund.
Conflict of Interest
None.
References
- Ohgaki H, Kleihues P (2005) Epidemiology and etiology of gliomas. Acta Neuropathol 109(1): 93-108.
- Maher EA, Brennan C, Wen PY, Durso L, Ligon KL, et al. (2006) Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res 66(23): 11502-11513.
- Cohen MH, Johnson JR, Pazdur R (2005) Food and Drug Administration Drug approval summary: temozolomide plus radiation therapy for the treatment of newly diagnosed glioblastoma multiforme. Clin Cancer Res 11(19 Pt 1): 6767-6771.
- Sun T, Warrington NM, Luo J, Brooks MD, Dahiya S, et al. (2014) Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J Clin Invest 124(9): 4123-4133.
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, et al. (2016) Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66(4): 271-289.
- Stupp R, Pavlidis N, Jelic S, Guidelines Task Force (2005) ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of malignant glioma. Ann Oncol 16(1) i64-i65.
- Schreck KC, Grossman SA (2018) Role of Temozolomide in the Treatment of Cancers Involving the Central Nervous System. Oncology (Williston Park) 32(11): 555-560.
- Nikolova T, Roos WP, Krämer OH, Strik HM, Kaina B (2017) Chloroethylating nitrosoureas in cancer therapy: DNA damage, repair and cell death signaling. Biochim Biophys Acta Rev Cancer 1868(1): 29-39.
- Ashby LS, Smith KA, Stea B (2016) Gliadel wafer implantation combined with standard radiotherapy and concurrent followed by adjuvant temozolomide for treatment of newly diagnosed high-grade glioma: a systematic literature review. World J Surg Oncol 14(1): 225.
- Champeaux C, Weller J (2020) Implantation of carmustine wafers (Gliadel®) for high-grade glioma treatment. A 9-year nationwide retrospective study. J Neurooncol 147(1): 159-169.
- Mao Z, Shen K, Zhu L, Xu M, Yu F, et al. (2019) Comparisons of cardiotoxicity and efficacy of anthracycline-based therapies in breast cancer: A network meta-analysis of randomized clinical trials. Oncol Res Treat 42(7-8): 405-413.
- Mao Z, Shen K, Zhu L, Xu M, Yu F, et al. (2019) Comparisons of cardiotoxicity and efficacy of anthracycline-based therapies in breast cancer: A network meta-analysis of randomized clinical trials. Oncol Res Treat 42(7-8): 405-413.
- Chamberlain FE, Jones RL, Chawla SP (2019) Aldoxorubicin in soft tissue sarcomas. Future Oncol 15(13): 1429-1435.
- Groves MD, Portnow J, Boulmay BC, Chawla SP, Dinh H, et al. (2016) Phase 2 study of aldoxorubicin in relapsed glioblastoma. Journal of Clinical Oncology 34(15 Suppl): 2027-2027.
- Carter TC, Medina-Flores R, Lawler BE (2018) Glioblastoma Treatment with Temozolomide and Bevacizumab and Overall Survival in a Rural Tertiary Healthcare Practice. Biomed Res Int 2018: 6204676.
- Seystahl K, Hentschel B, Loew S, Gramatzki D, Felsberg J, et al. (2020) Bevacizumab versus alkylating chemotherapy in recurrent glioblastoma. J Cancer Res Clin Oncol 146(3): 659-670.
- Schreck KC, Grossman SA (2018) Role of Temozolomide in the Treatment of Cancers Involving the Central Nervous System. Oncology (Williston Park) 32(11): 555-560.
- Zhang J, Stevens MF, Bradshaw TD (2012) Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol 5(1): 102-114.
- Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T (2007) Repair of alkylated DNA: recent advances. DNA Repair (Amst) 6(4): 429-442.
- D'Atri S, Tentori L, Lacal PM, Graziani G, Pagani, E et al. (1998) Involvement of the mismatch repair system in temozolomide induced apoptosis. Mol Pharmacol 54(2): 334-341.
- Aasland D, Götzinger L, Hauck L, Berte N, Meyer J, et al. (2019) Temozolomide Induces Senescence and Repression of DNA Repair Pathways in Glioblastoma Cells via Activation of ATR-CHK1, p21, and NF-κ Cancer Res 79(1): 99-113.
- Baer JC, Freeman AA, Newlands ES, Watson AJ, Rafferty JA, et al. (1993) Depletion of O6-alkylguanine-DNA alkyltransferase correlates with potentiation of temozolomide and CCNU toxicity in human tumour cells. Br J Cancer 67(6): 1299-1302.
- Kanzawa T, Bedwell J, Kondo Y, Kondo S, Germano IM (2003) Inhibition of DNA repair for sensitizing resistant glioma cells to temozolomide. J Neurosurg 99(6): 1047-1052.
- Agnihotri S, Gajadhar AS, Ternamian C, Gorlia T, Diefes KL, et al. (2012) Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest 122(1): 253-266.
- Loeb LA, Preston BD (1986) Mutagenesis by purinic/apyrimidinic sites. Annu Rev Genet 20: 201-230.
- Berdis AJ (2001) Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry 40(24): 7180-7191.
- Sabouri N, Johansson E (2009) Translesion synthesis of abasic sites by yeast DNA polymerase epsilon. J Biol Chem 284(46): 31555-31563.
- Wilson DM, Barsky D (2001) The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res 485(4): 283-307.
- Roos WP, Kaina B (2013) DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 332(2): 237-248.
- Quiros S, Roos WP, Kaina B (2010) Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell Cycle 9(1): 168-178.
- Hazra TK, Roy R, Biswas T, Grabowski DT, Pegg AE, et al. (1997) Specific recognition of O6-methylguanine in DNA by active site mutants of human O6-methylguanine-DNA methyltransferase. Biochemistry 36(19): 5769-5776.
- Gouws C, Pretorius PJ (2011) O6-methylguanine-DNA methyltransferase (MGMT): can function explain a suicidal mechanism? Med Hypotheses 77(5): 857-860.
- Dolan ME, Pegg AE, Dumenco LL, Moschel RC, Gerson SL (1991) Comparison of the inactivation of mammalian and bacterial O6-alkylguanine-DNA alkyltransferases by O6-benzylguanine and O6-methylguanine. Carcinogenesis 12(12): 2305-2309.
- Schold SC Jr, Kokkinakis DM, Rudy JL, Moschel RC, Pegg AE (1996) Treatment of human brain tumor xenografts with O6-benzyl-2'-deoxyguanosine and BCNU. Cancer Res 56(9): 2076-2081.
- Cabrini G, Fabbri E, Lo Nigro C, Dechecchi MC, Gambari R (2015) Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma. Int J Oncol 47(2): 417-428.
- Weller M, Tabatabai G, Kästner B, Felsberg J, Steinbach JP, et al. (2015) MGMT Promoter Methylation Is a Strong Prognostic Biomarker for Benefit from Dose-Intensified Temozolomide Rechallenge in Progressive Glioblastoma: The DIRECTOR Trial. Clin Cancer Res 21(9): 2057-2064.
- Binabaj MM, Bahrami A, ShahidSales S, Joodi M, Joudi Mashhad M, et al. (2018) The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J Cell Physiol 233(1): 378-386.
- Martinez, R, Schackert G, Yaya-Tur R, Rojas-Marcos I, Herman JG, et al. (2007) Frequent hypermethylation of the DNA repair gene MGMT in long-term survivors of glioblastoma multiforme. J Neurooncol 83(1): 91-93.
- Hegi ME, Liu L, Herman JG, Stupp R, Wick W, et al. (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol 26(25): 4189-4199.
- Sasmita AO, Wong YP, Ling APK (2018) Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol 14(1): 40-51.
- Rao AM, Quddusi A, Shamim MS (2018) The significance of MGMT methylation in Glioblastoma Multiforme prognosis. J Pak Med Assoc 68(7): 1137-1139.
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, et al. (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352(10): 997-1003.
- Morandi L, Franceschi E, de Biase D, Marucci G, Tosoni A, et al. (2010) Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer 10: 48.
- Kim DC, Kim KU, Kim YZ (2016) Prognostic Role of Methylation Status of the MGMT Promoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients. J Korean Neurosurg Soc 59(1): 26-36.
- Mansouri A, Hachem LD, Mansouri S, Nassiri F, Laperriere NJ, et al. (2019) MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol 21(2): 167-178.
- Thomas A, Tanaka M, Trepel J, Reinhold WC, Rajapakse VN, et al. (2017) Temozolomide in the Era of Precision Medicine. Cancer Res 77(4): 823-826.
- Baer JC, Freeman AA, Newlands ES, Watson AJ, Rafferty JA, et al. (1993) Depletion of O6-alkylguanine-DNA alkyltransferase correlates with potentiation of temozolomide and CCNU toxicity in human tumour cells. Br J Cancer 67(6): 1299-1302.
- Kanzawa T, Bedwell J, Kondo Y, Kondo S, Germano IM (2003) Inhibition of DNA repair for sensitizing resistant glioma cells to temozolomide. J Neurosurg 99(6): 1047-1052.
- Agnihotri S, Gajadhar AS, Ternamian C, Gorlia T, Diefes KL, et al. (2012) Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest 122(1): 253-266.
- Schold SC Jr, Kokkinakis DM, Chang SM, Berger MS, Hess KR, et al. (2004) O6-benzylguanine suppression of O6-alkylguanine-DNA alkyltransferase in anaplastic gliomas. Neuro Oncol 6(1): 28-32.
- Friedman HS, Dolan ME, Moschel RC, Pegg AE, Felker GM, et al. (2002) Enhancement of nitrosourea activity in medulloblastoma and glioblastoma multiforme. J Natl Cancer Inst 84(24): 1926-1931.
- Wedge SR, Newlands ES (1996) O6-benzylguanine enhances the sensitivity of a glioma xenograft with low O6-alkylguanine-DNA alkyltransferase activity to temozolomide and BCNU. Br J Cancer 73(9): 1049-1052.
- Quinn JA, Jiang SX, Carter J, Reardon DA, Desjardins A, et al. (2009) Phase II trial of Gliadel plus O6-benzylguanine in adults with recurrent glioblastoma multiforme. Clin Cancer Res 15(3): 1064-1068.
- Kunkel TA, Erie DA (2005) DNA mismatch repair. Annu Rev Biochem 74: 681-710.
- Lenhart JS, Pillon MC, Guarné A, Biteen JS, Simmons LA (2016) Mismatch repair in Gram-positive bacteria. Res Microbiol 167(1): 4-12.
- Schofield MJ, Hsieh P (2003) DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol 57: 579-608.
- Guarné A, Charbonnier JB (2015) Insights from a decade of biophysical studies on MutL: Roles in strand discrimination and mismatch removal. Prog Biophys Mol Biol 117(2-3): 149-156.
- Masih PJ, Kunnev D, Melendy T (2008) Mismatch Repair proteins are recruited to replicating DNA through interaction with Proliferating Cell Nuclear Antigen (PCNA). Nucleic Acids Res 36(1): 67-75.
- Surtees JA, Alani E (2004) Replication factors license exonuclease I in mismatch repair. Mol Cell 15(2): 164-166.
- Lancey C, Tehseen M, Raducanu VS, Rashid F, Merino N, et al. (2020) Structure of the processive human Pol δ Nat Commun 11(1): 1109.
- Bowen N, Kolodner RD (2017) Reconstitution of Saccharomyces cerevisiae DNA polymerase ε-dependent mismatch repair with purified proteins. Proc Natl Acad Sci USA 114: 3607-3612.
- Yi GZ, Huang G, Guo M, Zhang X, Wang H, et al. (2019) Acquired temozolomide resistance in MGMT-deficient glioblastoma cells is associated with regulation of DNA repair by DHC2. Brain 142(8): 2352-2366.
- Perazzoli G, Prados J, Ortiz R, Caba O, Cabeza L, et al. (2015) Temozolomide Resistance in Glioblastoma Cell Lines: Implication of MGMT, MMR, P-Glycoprotein and CD133 Expression. PLoS One 10(10): e0140131.
- Friedman HS, McLendon RE, Kerby T, Dugan M, Bigner SH, et al. (1998) DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis andresponse to Temodal in newly diagnosed malignant glioma. J Clin Oncol 16(2): 3851-3857.
- Stark AM, Doukas A, Hugo HH, Hedderich J, Hattermann K, et al. (2015) Expression of DNA mismatch repair proteins MLH1, MSH2, and MSH6 in recurrent glioblastoma. Neurol Res 37(2): 95-105.
- Indraccolo S, Lombardi G, Fassan M, Pasqualini L, Giunco S, et al. (2019) Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clin Cancer Res 25(6): 1828-1837.
- Struve N, Binder ZA, Stead LF, Brend T, Bagley SJ, et al. (2020) EGFRvIII upregulates DNA mismatch repair resulting in increased temozolomide sensitivity of MGMT promoter methylated glioblastoma. Oncogene 39(15): 3041-3055.
- Nagel ZD, Kitange GJ, Gupta SK, Joughin BA, Chaim IA, et al. (2017) DNA Repair Capacity in Multiple Pathways Predicts Chemoresistance in Glioblastoma Multiforme. Cancer Res 77(1): 198-206.
- Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T (2007) Repair of alkylated DNA: recent advances. DNA Repair (Amst) 6(4): 429-442.
- Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, et al. (2008) Eukaryotic Y-family polymerases bypass a 3-methyl-2'-deoxyadenosine analog in vitro and methyl methane sulfonate-induced DNA damage in vivo. Nucleic Acids Res 36(7): 2152-2162.
- Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M (2009) ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet 5(1): e1000324.
- Loeb LA, Preston BD, Snow ET, Schaaper RM (1986) Apurinic sites as common intermediates in mutagenesis. Basic Life Sci 38: 341-347.
- Rubinson EH, Christov PP, Eichman BF (2013) Depurination of N7-methylguanine by DNA glycosylase AlkD is dependent on the DNA backbone. Biochemistry 52(42): 7363-7365.
- Taylor JS (2002) New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat Res 510(1-2): 55-70.
- Sabouri N, Johansson E (2009) Translesion synthesis of abasic sites by yeast DNA polymerase epsilon. J Biol Chem 284(46): 31555-31563.
- Yang Z, Price NE, Johnson KM, Wang Y, Gates KS (2017) Interstrand cross-links arising from strand breaks at true abasic sites in duplex DNA. Nucleic Acids Res 45(11): 6275-6283.
- Robertson AB, Klungland A, Rognes T, Leiros I (2009) DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci 66(6): 981-993.
- Lindahl T (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res 462(2-3): 129-35.
- Goellner EM, Grimme B, Brown AR, Lin YC, Wang XH, et al. (2011) Overcoming temozolomide resistance in lioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Res 71(6): 2308-2317.
- Shi J, Dong B, Zhou P, Guan W, Peng Y (2017) Functional network analysis of gene phenotype connectivity associated with temozolomide. Oncotarget 8(50): 87554-87567.
- Bobola MS, Kolstoe DD, Blank A, Chamberlain MC, Silber JR (2012) Repair of 3-methyladenine and abasic sites by base excision repair mediates glioblastoma resistance to temozolomide. Front Oncol 2: 176.
- Higuchi F, Nagashima H, Ning J, Koerner MVA, Wakimoto H, et al. (2020) Restoration of Temozolomide Sensitivity by PARP Inhibitors in Mismatch Repair Deficient Glioblastoma is Independent of Base Excision Repair. Clin Cancer Res 26(7): 1690-1699.
- Tang JB, Svilar D, Trivedi RN, Wang XH, Goellner EM, et al. (2011) N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol 13(5): 471-486.
- Agnihotri S, Burrell K, Buczkowicz P, Remke M, Golbourn B, et al. (2014) ATM regulates 3-methylpurine-DNA glycosylase and promotes therapeutic resistance to alkylating agents. Cancer Discov 4(10): 1198-1213.
- Thanasupawat T, Natarajan S, Rommel A, Glogowska A, Bergen H, et al. (2017) Dovitinib enhances temozolomide efficacy in glioblastoma cells. Mol Oncol 11(8): 1078-1098.
- Parsons JL, Dianova I, Allinson, SL, Dianov GL (2005) Poly (ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J 272(8): 2012-2021.
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, et al. (2014) Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343(6167): 189-193.
- Reha-Krantz LJ, Nonay RL, Day RS, Wilson SH (1996) Replication of O6-methylguanine-containing DNA by repair and replicative DNA polymerases. J Biol Chem 271(33): 20088-20095.
- Chavarria D, Ramos-Serrano A, Hirao I, Berdis AJ (2011) Exploring the roles of nucleobase desolvation and shape complementarity during the misreplication of O(6)-methylguanine. J Mol Biol 412(3): 325-339.
- Jałoszyński P, Ohashi E, Ohmori H, Nishimura S (2005) Error-prone and inefficient replication across 8-hydroxyguanine (8-oxoguanine) in human and mouse ras gene fragments by DNA polymerase kappa. Genes Cells 10(6): 543-550.
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juárez R, Bebenek K, et al. (2005) A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell 19(3): 357-366.
- Bertocci B, De Smet A, Weill JC, Reynaud CA (2006) Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 25(1): 31-41.
- Wang H, Wu W, Wang HW, Wang S, Chen Y, et al. (2010) Analysis of specialized DNA polymerases expression in human gliomas: association with prognostic significance. Neuro Oncol 12(7): 679-686.
- Peng C, Chen Z, Wang S, Wang HW, Qiu W, et al. (2015) The Error-Prone DNA Polymerase κ Promotes Temozolomide Resistance in Glioblastoma through Rad17-Dependent Activation of ATR-Chk1 Signaling. Cancer Res 76(8): 2340-2353.
- Bostian AC, Maddukuri L, Reed MR, Savenka T, Hartman JH, et al. (2016) Kynurenine Signaling Increases DNA Polymerase Kappa Expression and Promotes Genomic Instability in Glioblastoma Cells. Chem Res Toxicol 29(1): 101-108.
- Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, et al. (2017) Comprehensive Analysis of Hypermutation in Human Cancer. Cell 171(5): 1042-1056.
- Choi JS, Kim S, Motea E, Berdis A (2017) Inhibiting translesion DNA synthesis as an approach to combat drug resistance to DNA damaging agents. Oncotarget 8(25): 40804-40816.
- Reineks EZ, Berdis AJ (2004) Evaluating the contribution of base stacking during trans lesion DNA replication. Biochemistry 43(2): 393-404.
- Choi JS, Kim CS, Berdis A (2018) Inhibition of Translesion DNA Synthesis as a Novel Therapeutic Strategy to Treat Brain Cancer. Cancer Res 78(4): 1083-1096.
-
Anthony Berdis. Temozolomide Resistance in Glioblastoma Multiforme: Mechanisms, Ramifications, and Potential Solutions to an Urgent Clinical Problem. On J Complement & Alt Med. 6(5): 2021. OJCAM.MS.ID.000647.
-
Temozolomide, Glioblastoma Multiforme, DNA-alkylating agent, Anti-cancer agent, TMZ-induced drug resistance, Glioblastoma multiforme, Translesion DNA synthesis, Nervous system, Ionizing radiation, Radiotherapy, DNA replications
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






