 Research Article
Research Article
Categorizing Molecular Features of Venom Toxins Using Bioinformatics Tools
Vinod P Sinoorkar1, Pratiksha D Shinde2*, Mohammed Danish A Shaikh3, Gouri S Mandrup4, Isha A Puranik5
Walchand Centre for Biotechnology, Solapur, Maharashtra, India
Pratiksha D Shinde, Walchand Centre for Biotechnology, Solapur, Maharashtra, India.
Received Date:March 08, 2023; Published Date:May 03, 2023
Abstract
Poisonous organisms are represented in many taxa, including kingdom Animalia. During evolution, animals have developed special organs for production and injection of venoms. Animal venoms are complex mixtures, compositions of which depend on species producing venom. The most known and studied poisonous terrestrial animals are snakes, scorpions and snails. Venomous animals produce a myriad of important pharmacological components. The individual components, or venoms (toxins), are used in ion channel and receptor studies, drug discovery, and formulation of insecticides. Knowing the key clinical applications of venom, the present investigation was carried out to understand the molecular basis of venom toxins of animals like snake (L-amino acid oxidases), cone snail (Contulakin-G) and scorpion (Chlorotoxin) by retrieving the protein sequence information, deducing various physicochemical properties, predicting secondary structural elements, homology modelling and depicting the potent antigenic regions using various bioinformatics tools and soft-wares. Because of their remarkable molecular diversity, venoms are key, albeit challenging, resource for pharmacological discovery that contribute to the development of drugs that act as anti-tumor agents, heart stimulants and therapies for neurological diseases [1-3]. Venom-informatics is a systematic bioinformatics approach in which classified, consolidated and cleaned venom data are stored into repositories and integrated with advanced bioinformatics tools for the analysis of structure and function of toxins. Venom-informatics complements experimental studies and helps reduce the number of essential experiments.
Keywords:Venom; Toxins; Venom-informatics; Therapeutics; snake; cone snail; Scorpion etc
Introduction
Venomous organisms are widespread throughout the globe and represented in many biological taxa [5]. Venom is a complex mixture of proteins and peptides and presents several medical and pharmaceutical applications. Over the years, venoms and fractions there, have been displayed several biological activities/ applications, including antibacterial, anti-protozoarian, antiviral (human immunodeficiency virus), analgesic, treatment of cancer and for many such diseases [6]. It is important to note that some of those aforementioned activities can be related not only to medium to high abundance specific venom toxins but also to low abundance components and, eventually, to their synergistic effects.
Venomous animals permanently or periodically contain toxins in their organisms, which are toxic to other species. Even small doses of such compounds in the body of another animal cause painful disorders and sometimes death. The pathophysiological complications related to a single sting of such animals are noteworthy to recognize venomous animal envenomation as a universal health problem [4]. However, the information about peptides and proteins from toxins is not standardized in these resources, especially the naming of toxins and pharmacological activities, and mining for venom peptides is difficult. By molecular cloning and classical biochemistry, several proteins and peptides (related to toxins) are characterized. The revelation of many other novel components and their potential activities in different fields of biological and medicinal sciences revitalized the interests in the field of animal venomics and venom-informatics [5].
Venom have been of great interest to researchers due to their significant roles in pharmacology [7]. In our current investigation we have studied three different toxins from three different venomous organisms namely L- amino acid oxidases (snake), Contulakin-G (cone snail), Chlorotoxin (scorpion). Snake venoms are sources of molecules with proven and potential therapeutic applications. However, it is still challenging to associate signaling pathways identified through functional genomics to the pathophysiology of snakebite (assessed through well-established biochemical and biological assays, screening for hemorrhagic, hypotensive, edematogenic, neurotoxic, and myotoxic activities) [8]. The geographic cone is the most toxic of the known species, and several human deaths have resulted from envenomation. Severe cases of cone snail stings involve muscle paralysis, blurred/double vision, and respiratory paralysis, leading to death. The animals produce a potent venom to paralyse their prey. The venom contains a complex mixture of substances that includes neurotoxins, which are chemicals that block the conduction of nerve impulses [9,10]. Scorpion’s venom is primarily considered as the cocktail of chemicals which are injected into the prey/predators to disrupt their biological systems. Chlorotoxin possesses targeting properties towards cancer cells including glioma, melanoma, small cell lung carcinoma, neuroblastoma and medulloblastoma [11].
Methodology
Retrieval of sequences
Protein sequence data of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) have been retrieved by UniprotKB database and saved in FASTA format with their respective UniprotKB IDs [12].
Analysis of physico-chemical properties
Physico-chemical properties of L-amino acid oxidases (snake), contulakin-G peptide (cone snail) and chlorotoxin (scorpion) were analysed by using Protparam tool available at Expasy server which included isoelectrical point, theoretical pI, extension coefficient, half-life, instability index, Aliphatic index, Grand average, hydropathicity, etc [13].
Secondary structure prediction
The secondary structures of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were predicted by SOPMA, which gives the information of Alpha helix, beta sheets, extended strands and random coils [14].
Tertiary structure prediction
The tertiary structures of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were obtained by using SWISS-MODEL tool and PDBsum by selecting the template with maximum homology and with optimized parameters. The obtained structures were stored in pdb format for visualization.
Visualization of tertiary structures
The predicted tertiary structures of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were visualized by using structure visualization tool Rasmol. Visualization was done using different models and formats to understand structural features of respected proteins.
Antigenicity Prediction
The antigenicity of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were obtained by using Antigenicity prediction tool. Kolaskar and Tongaonkar predict the segment within a protein sequence that are likely to be antigenic by eliciting an antibody response [15,16].
Prediction of helical wheel projection
The properties of alpha helices in proteins were obtained by a helical wheel projection tool, which is a type of plot or visual representation. The amino acids sequence that makes up a helical region of the protein’s secondary structure are plotted in a rotating manner where the angle of rotation between consecutive amino acids is 100°, so that the final representation looks down the helical axis.
Results and Discussion
Retrieval of sequences
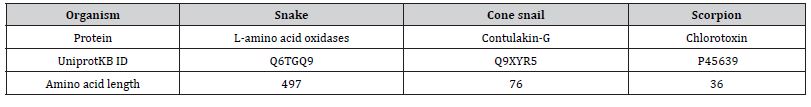
The sequences of L-amino acid oxidases (snake contulakin-G (cone snail) and chlorotoxin (scorpion) were retrieved from Uniprot KB. The sequences obtained were stored in FASTA format with their respective UniprotKB IDs. The UniprotKB IDs, sequence length and protein names are shown (Table 1).
Table 1: Retrieval of protein sequences.
Analysis of Physico-Chemical Properties
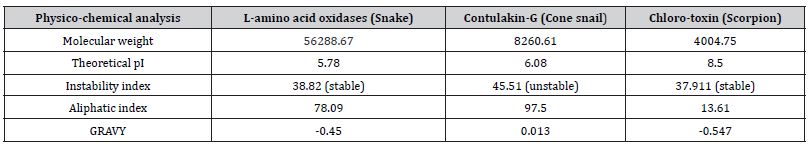
The analysis of physicochemical properties of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) was done by using Protparam tool and respective results were shown (Table 2). The theoretical pI depicts that chlorotoxin (scorpion) is basic in nature, while L-amino acid oxidases (snake) and contulakin-G (cone snail) both are acidic. Further, the instability index indicates that contulakin-G (cone snail) is unstable but L-amino acid oxidases (snake) and chlorotoxin (scorpion) are stable.
Table 2: Physicochemical analysis.
Secondary structure prediction
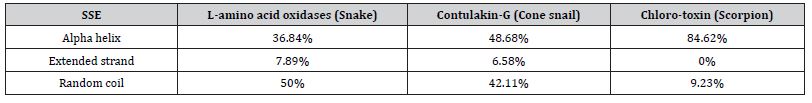
The secondary structures of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were predicted by SOPMA tool. The secondary structure elements alpha helix, extended strand and random coils were calculated shown (Table 3). According to Table 3 the percentage of alpha helix was found to be more in chlorotoxin (scorpion) and least in L-amino acid oxidases (snake). Further, the percentage of extended strand was found to be more in L-amino acid oxidases (snake) and least in chlorotoxin (scorpion).
Table 3: Secondary structure prediction.
Homology Modelling
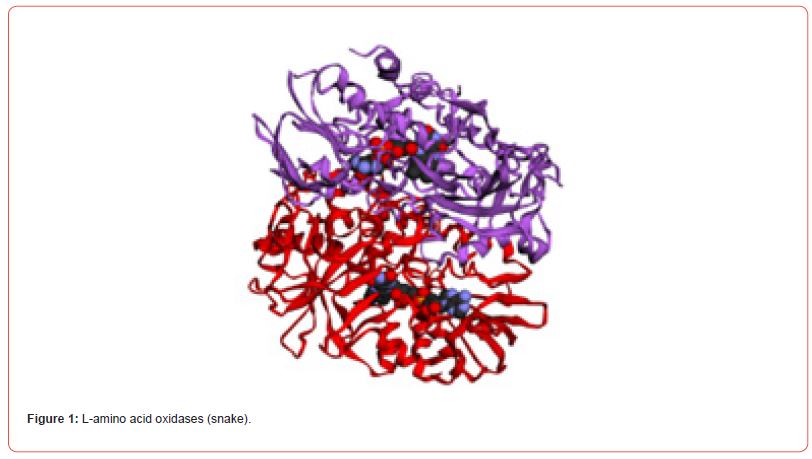
The 3D structures of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were predicted by using automated mode of SWISS-MODEL server.
Visualization of tertiary structures
The predicted tertiary structures of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were visualized by using structure visualization tool RasMol. The generated structures are shown (Figure 1-6).
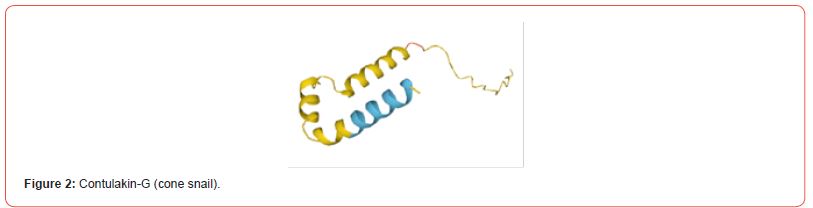
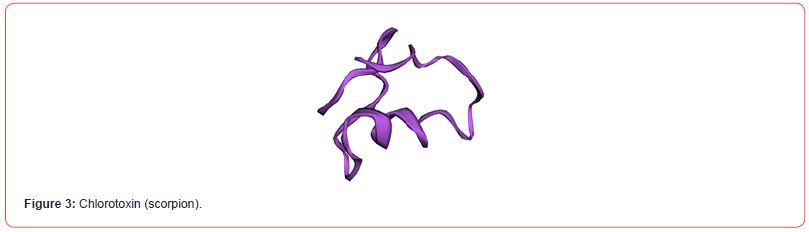
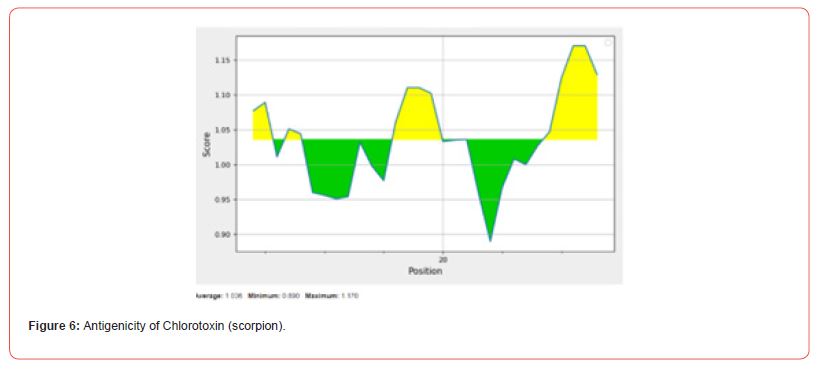
Antigenicity Prediction
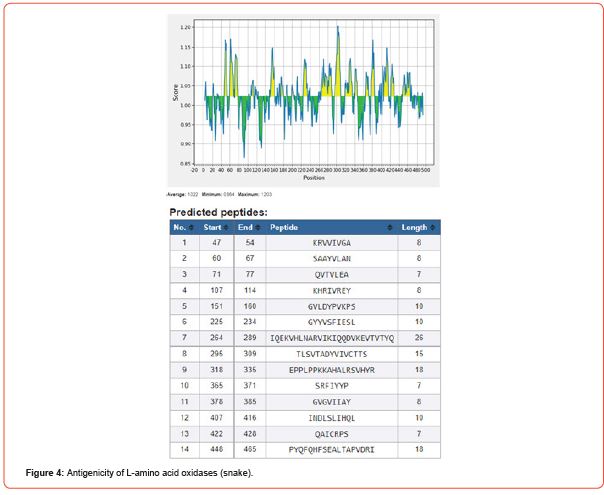
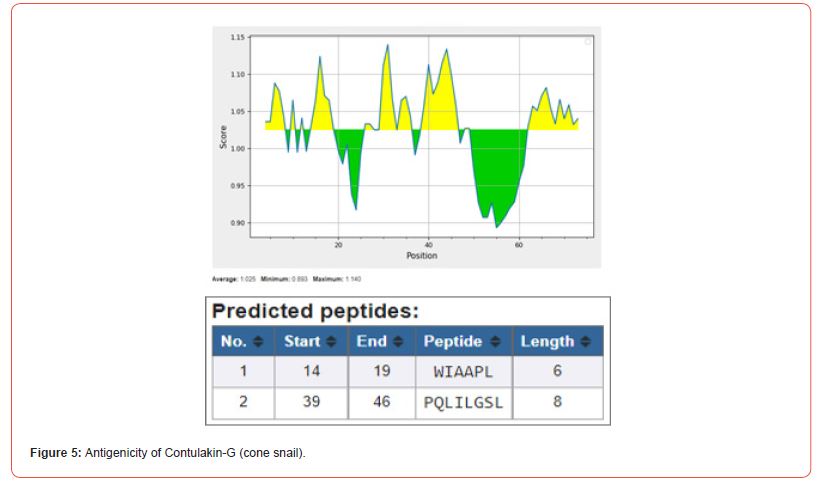
Kolaskar and Tongaonkar’s method predicts antigenic epitopes of given sequence, based on physicochemical properties of amino acid residues that frequently occur in experimentally determined antigenic epitopes. In-silico studies reveals that L-amino acid oxidase with 14 antigenic peptides is found to be more antigenic while contulakin-G with 2 and chlorotoxin with 1 antigenic peptide being lower antigenic nature [17].
Prediction of helical wheel projection
Helical wheels are a standard way to predict protein sequence segments with either helical or non-helical potential. These diagrams are projections of a protein’s amino acid sequence onto a plane perpendicular to the axis of the helix. Comparing helical wheel representations for helical segments and non-helical segments made it clear that residues with hydrophobic side chains occur significantly more often in helical regions. One the other hand, the non-helical regions have more polar side chains [18,19].

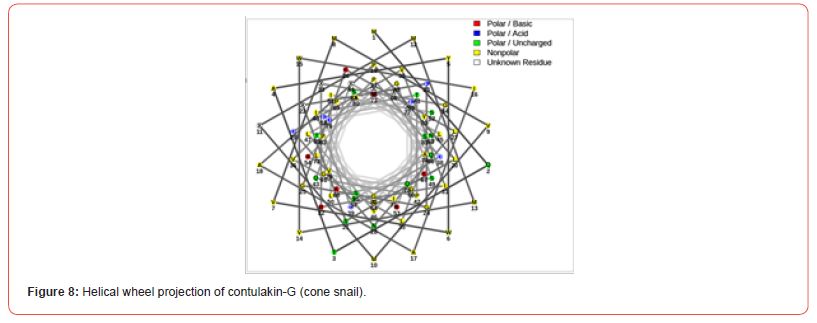
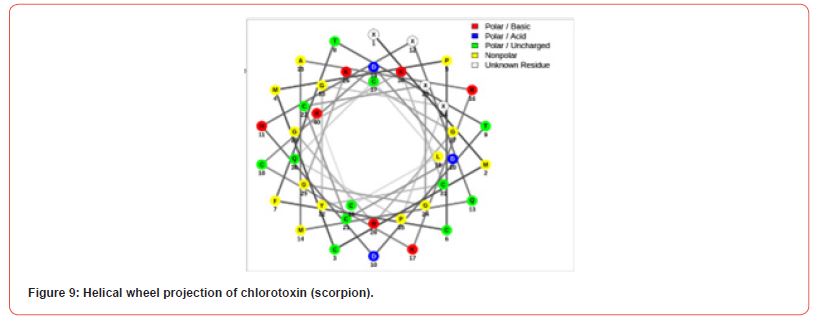
Conclusion
Proteins of three different venomous organisms were chosen for the investigation namely L-amino acid oxidases (snake), Contulakin-G (cone snail), Chlorotoxin (scorpion). In the present study, sequence and structural insight of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) emphasize their basic molecular nature with knowledge of venom toxin composition and their biological properties for designing novel scaffold and designing new peptide-based drugs for the treatment of various cancers, haemostatic and other chronic disorders and as well as new tools for clinical diagnostic and assays of haemostatic parameters. Sequences of L-amino acid oxidases (snake), contulakin-G (cone snail) and chlorotoxin (scorpion) were retrieved from UniProtKB protein database, using these sequences their physicochemical properties were analysed from Protparam tool. Further, secondary structural elements were predicted from SOPMA tool and the tertiary models of respected proteins were built using SWISS-MODEL server. Additionally, antigenicity of respected proteins was predicted using kolaskar and tongaonkar’s antigenicity prediction tool along with prediction of helical wheel projection.
Acknowledgement
None.
Conflicts of Interest
No conflict of interest.
- Liu L, Chen Q, Yang J, Gang W, Zhao L, et al. (2022) Fire Needling Acupuncture for Adult Patients with Acute Herpes Zoster: Protocol of a Systematic Review and Meta-Analysis. J Pain Res 15: 2161-2170.
- Wu L, Chen Y, Man T, Jin L, Xu Z, et al. (2022) External Therapy of Chinese Medicine for Herpes Zoster: A Systematic Review and Meta-Analysis. Evid Based Complement Alternat Med 2022: 3487579.
- Pei W, Zeng J, Lu L, Lin G, Ruan J (2019) Is acupuncture an effective postherpetic neuralgia treatment? A systematic review and meta-analysis. J Pain Res 12: 2155-2165.
- Bian Z, Yu J, Tu M, Liao B, Huang J, et al. (2022) Acupuncture therapies for postherpetic neuralgia: a protocol for a systematic review and Bayesian network meta-analysis. BMJ Open 12(3): e056632.
- Wang H, Wan R, Chen S, Qin H, Cao W, et al. (2022) Comparison of Efficacy of Acupuncture-Related Therapy in the Treatment of Postherpetic Neuralgia: A Network Meta-Analysis of Randomized Controlled Trials. Evid Based Complement Alternat Med 2022: 3975389.
- Münstedt K (2019) Bee products and the treatment of blister-like lesions around the mouth, skin and genitalia caused by herpes viruses-A systematic review. Complement Ther Med 43: 81-84.
- Rocha MP, Amorim JM, Lima WG, Brito JCM, da Cruz Nizer WS (2021) Effect of honey and propolis, compared to acyclovir, against Herpes Simplex Virus (HSV)-induced lesions: A systematic review and meta-analysis. J Ethnopharmacol 287: 114939.
- Yosri N, Abd El-Wahed AA, Ghonaim R, Khattab OM, Sabry A, et al. (2021) Anti-Viral and Immunomodulatory Properties of Propolis: Chemical Diversity, Pharmacological Properties, Preclinical and Clinical Applications, and In Silico Potential against SARS-CoV-2. Foods 10(8): 1776.
- Zeng JC, Zhang RL, Wei XJ, Lin GH (2022) Acupuncture for improving a case of widespread herpes zoster after non-Hodgkin's lymphoma chemotherapy. Explore (NY) 18(5): 608-611.
- Jiang Y, Zheng RX, Yu ZY, Zhang XW, Li J, et al. (2022) Traditional Chinese Medicine for HIV-Associated Acute Herpes Zoster: A Systematic Review and Meta-Analysis of Randomized Trials. Evid Based Complement Alternat Med 2022: 8674648.
-
Vinod P Sinoorkar, Pratiksha D Shinde*, Mohammed Danish A Shaikh, Gouri S Mandrup, Isha A Puranik. Categorizing Molecular Features of Venom Toxins Using Bioinformatics Tools. On J Complement & Alt Med. 8(3): 2023. OJCAM.MS.ID.000689.
-
Venom; Toxins; Venom-informatics; Therapeutics; Snake; Cone snail; Scorpion; Bioinformatics; Venomous animals; Painful disorders; Pathophysiological complications
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






