 Research Article
Research Article
Nasopharyngeal Colonization with Streptococcus Pneumoniae in Healthy Infants Less Than 60 Days Attending the Outpatient Clinic of Alexandria University Children’s Hospital
Ahmed El Nawawy, Soad Farid Hafez, Manal Abdel Malek Antonios and Ashraf Khamis Mahmoud*
Department of pediatrics, Faculty of Medicine, Alexandria University, Egypt
Ashraf Khamis Mahmoud, Department of pediatrics, Faculty of Medicine, Alexandria University, Egypt.
Received Date: November 06, 2019; Published Date: November 14, 2019
Abstract
Streptococcus pneumoniae (pneumococcus) is a bacterial pathogen that affects children and adults worldwide [1]. Although S. pneumoniae is the leading bacterial cause of respiratory tract infections, it is also capable of causing a wide variety of infectious syndromes including meningitis, peritonitis, and sepsis [2-4]. The present study is to determine the prevalence of pneumococcus colonization among apparently healthy infants below 60 days of age and to identify the prevalent pneumococcal serotypes among this infant population In this study over a period of 14 months. Sample size was found to be 531 infants. The sample was selected by the systematic random procedure [5]. The results revealed that 0.75 % (n=4/531) of infants showed positive carriage of pneumococcus in their nasopharynx.
After serotyping, only one case of 4 strept. Pneumonia identified as vaccine included type and the remaining 3 were of non –vaccine type and this result was checked by real-time PCR serogrouping.75% of cases were sensitive to penicillin and the remaining 25% were resistant.
Keywords: Pneumococcal carriage; Healthy infants; Prevalence in less than 60 days; Pneumococcal serotypes; Penicillin sensitivity
Introduction
Streptococcus pneumoniae (pneumococcus) is a bacterial pathogen that affects children and adults worldwide [1]. Although S. pneumoniae is the leading bacterial cause of respiratory tract infections, it is also capable of causing a wide variety of infectious syndromes including meningitis, peritonitis, and sepsis [2-4]. Virulence factors include capsule, pnemolysin, the Pneumococcal Pilus, Choline-Binding Proteins (CBPs) and pneumococcal surface proteins and other surface proteins [6].
Nasopharyngeal colonization usually is acquired around 6 months of age but can occur as early as the first weeks of life, and although the average length of carriage is 3 to 4 months, some individuals (especially young infants) may harbor pneumococci for over a year [7].
Nasopharyngeal colonization S. pneumoniae in children mainly depends on age.
Socioeconomic and environmental risk factors include family size, income, smoking and antibiotic use [8-10].
The nasopharyngeal flora become established during the first months of life. A broad variety of microorganisms including S. pneumoniae, H influenzae, and M catarrhalis [8].
Colonization not only is obligatory for invasive disease, but also provides the basis for horizontal spread of pneumococci [11].
The serotype distributing among nasopharyngeal carriage isolates varies slightly by country, age-group.
Based on the results of these clinical trials, in 2000, ACIP recommended routine use of PCV7 for all children aged 2-59 month [12].
The 13-valent pneumococcal conjugate vaccine (PCV13) contains serotypes 1, 3,4, 5, 6A,6B, 7F,9V, 14, 18C, 19A, 19F, and 23F.
Aim of the Work
The aim of the present study is to determine the prevalence of pneumococcus colonization among apparently healthy infants below 60 days of age attending the outpatient clinic of Elshatby University children’s hospital, to identify the prevalent pneumococcal serotypes among this infant population and to determine the resistance to penicillin among the isolated pneumococci.
Subjects
This prospective study conducted over a period of 14 month from 1st of Feb-2018 to 31st of Apr-2019. Healthy cases below the age of 60 days presented to the outpatient clinic of Alexandria university children’s hospital enrolled in this study.
Prior to enrollment of any child in the study and sample collection an informed consent was obtained from his parents or guardians.
In this study over a period of 14 months. By using a power of 80%, precision 5%, α=0.05. Sample size was found to be 531 infants.
The sample was selected by the systematic random procedure.
The following criteria are excluded Any infant with clinical evidence of infection (such as fever, respiratory infection, sepsis, meningitis, otitis media, ….etc.), Any infant indicated for admission to hospital or was previously hospitalized, Any infant with serological evidence of infection (elevated C-reactive proteinprocalcitonin- leukocytosis-any acute phase reactants positive)and Any infant who received antibiotic in the previous 60 days.
Methods
All enrolled infants subjected to the following:
Clinical data included
Thorough history taking and clinical examination including age, sex, feeding history (formula or breast),labor, anthropometric measures, weight, height.
Sample collection
Collection of Nasopharyngeal swabs.
a) A flexible fine shafted swab flexible swab was used.
b) The distance from the infant’s nose to the ear used to give an estimate of the distance the swab should be inserted.
c) The swab inserted into one nostril straight back (not upwards) and back to the nasopharynx.The swab rotated 2-3 times and held swab in place for 5 seconds to ensure maximum absorbency. The swab then be slowly withdrawn with a rotating motion.The procedure was repeated with the same swab for the second nostril to deliver a combined sample.
d) All nasopharyngeal swabs immediately inoculated on (gentamicin blood agar).
Then inoculated in 1ml Skim Milk-Tryptone-Glucose-Glycerol (STGG) medium which was properly shacked/vortexed prior to sample collection.
Inoculated STGG medium labeled and transported to the lab. In the lab they will be vortexed and subsequently frozen at -20°C/- 70°C.
Throat Swab
e) A wooden tongue depressor used to hold the tongue in place.
f) Without touching the sides of the mouth, A sterile swab used to swab the posterior nasopharynx.
g) All throat swabs immediately inoculated on (gentamicin blood agar).
h) Then inserted into STGG medium.
Laboratory assessment (bacteriological investigations)
a) Isolation of S. pneumoniae from the inoculated STGG vials: Immediately vortexed and stored at -20°C until appropriate sample processing is feasible.
Thawed STGG vials will be vortexed then 200μl will be inoculated into 5ml enriched Todd-Hewitt with 0.5% Yeast and 1ml rabbit serum and incubated for 4-6 hours at 37°C in 5% CO2 atmosphere.
Subsequently, 10μl volumetric loop will be used to inoculate gentamicin blood agar plates for selective isolation of S. pneumoniae [13].
b) Identification of S. pneumoniae: Identification of S. pneumoniae were based on colonial morphology, microscopic morphology, Gram staining reaction, optochin sensitivity and bile solubility. All identified pure cultures of isolates of S. pneumonia were inoculated in 1ml STGG medium and stored at -20°C. All isolates were subjected to full microbiological identification based on conventional microbiological techniques and antimicrobial sensitivity testing based on the standards set by Clinical Laboratory Standards Institute (CLSI) for antimicrobial susceptibility testing.
c) Serotyping of S. pneumoniae: All S. pneumoniae isolates will be subjected to serotyping using capsular Quellung reaction.
The Pneumococcal serotypes are 95, so, we chose all groups of pneumococcal serotypes in which all PCV13 serotypes were present (as these serotypes were claimed to be the most pathogenic serotypes all over the world).
a) The Pneumococcal groups used were:
Group A which contains serotypes (1, 2, 4, 5, 18F, 18A, 18B and 18C).
Group B which contains serotypes (3,6A, 6B, 6C, 8, 19F, 19A, 19B and 19C).
Group H which contains serotypes (13, 14, 15F, 15B, 15C, 23F, 23A, 23B, 28F and 28A).
Group P which contains serotypes (1,7F, 7A, 7B, 7C, 14, 19F, 19A, 19B, and,19C).
Group R which contains serotypes (3, 4, 9A, 9L, 9N, 9V, 12F, 12A, and 12G).
Those who don’t belong to any of the above groups, belong to other groups which were isolated but not serotyped and assigned as non-vaccine serotypes (NVSTs)
After grouping, serotyping of the 13 VSTs (vaccine serotypes) were performed using Quellung reaction.
Isolates were tested using grouped antisera(mentioned above) followed by specific tests for the 13 serotypes included in PCV13. The positive Pneumococcal isolates that weren’t included in PCV13 were considered positive for pneumococci but not typable (nonvaccine serotypes, NVSTs).
Isolates of S. pneumoniae from infants were challenged against antisera from Staten’s Serum Institute, Copenhagen, Denmark against the 13 valency pneumococcal vaccine serotypes.
d) Screening for pneumococcal carriage:
The CDC protocol for: “Identification of pneumococcal serotypes from nasopharynx using culture and PCR” was adopted.
Confirmatory test to differentiate S. pneumoniae from other oral viridians streptococci was accomplished by detection of:
Optochin susceptibility
Bile solubility
e) Molecular typing by multiplex PCR
All pneumococcal isolates used in the PCR assays were delinked with the results of serotyping by quelling reaction, and the researcher who performed the PCR was also blinded to these results.
DNA Extraction was done by suspending 3-5 colonies of fresh overnight single colony subculture on blood agar in 50μl 0.02 M NaOH / 0.1% SDS (Sodium dodecyl sulphate) followed by heating at 95°C for 15 min. Then the suspension was diluted with 10-fold sterile distilled water, Centrifuged at 9000 rpm for 5 min and 5μL of the supernatant was used as a template in the PCR reaction.
A set of 13 primer pairs were used, one primer (cpsA) served as internal control. It targets cpsA, a highly conserved gene that exists in all capsular loci that have been characterized till now. The other 12 primer pairs designed to target genes specific for vaccine covered serotypes 1, 3, 4, 5, 6A/6B/6C, 7F/7A, 9V/9A 14, 18C, 19F, 19A, and 23F (Invitrogen by life technologies, Thermo Fisher Scientific Inc. USA). (Table 1)
Three multiplex reactions were sequentially performed.
The PCRs were performed in 25μL volumes, with each reaction mixture containing the following: Maxima Hot Start Green PCR Master Mix (Thermo Fisher Scientific Inc. USA) 12.5ul, Primers with pre-determined concentrations, 5μL of the DNA extract and distilled nuclease free water to a final volume of 25ul. The amplification reactions were carried out using thermal cycler (Thermal cycler 2720, Applied Biosystems, USA) with the following cycling conditions: 94°C for 4 min, followed by 30 cycles of 94°C for 45 sec, 54°C for 45 sec and 72°C for 2 min, then final extension of 72°C for 2 min.
Each PCR included distilled water as a negative control and DNA extracted from pneumococcal isolates from which serotypes were previously determined by the quellung reaction as a positive control.
PCR products were electrophoresed on 1.5% agarose gel stained with ethidium bromide (Sigma, USA) and visualized under UV light. The molecular sizes of the amplicons were determined by comparison to a 100 bp DNA ladder [14].
Results
The results revealed that 0.75 % (n=4/531) of infants showed positive carriage of pneumococcus in their nasopharynx. After serotyping, only one case of 4 strept. Pneumonia identified as vaccine included type and the remaining 3 were of non –vaccine type and this result was checked by real-time PCR serogrouping. 75% of cases were sensitive to penicillin and the remaining 25% were resistant (Table 1).
Table 1: Demographic characteristics of studied population.
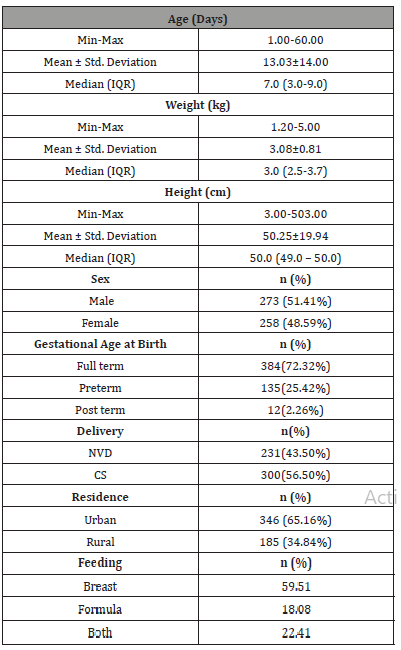
The total recruited cases (n=531) randomly selected after verification of exclusion criteria had a median age of 7 days with a minimum of 1 day and a maximum age of 60 days. Their weight ranged from 1.2-5 kg with a median of 3 kg .The studied population were of standard height, the mean height was 50.25 cm. Both sexes were included with 51.41% males and 48.59% females, most of them were full term (72.32%). Caesarian section exceeded normal vaginal delivery (56.5% compared to 43.5% respectively). Most of the studied cases were from urban origin (65.16%).
Regarding their feeding, exclusive breast feeding was reported in 59.51% of cases, 22.41% received mixed formula and breast feeding while the remaining 18% received formula feeding solely (Table 2).
Table 2: Shows the rate of recruitment of studied cases.
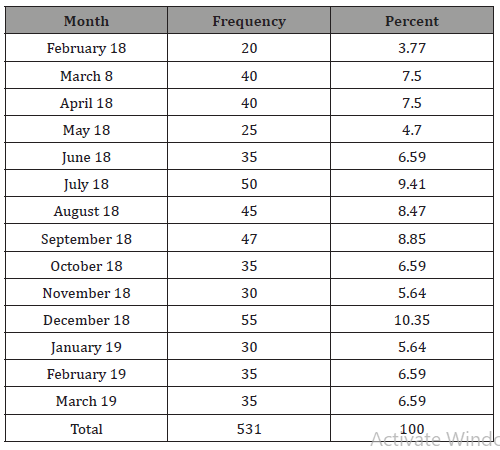
Table 3: Relation between strept. Pneumonia carriage and different risk factors of studied population.
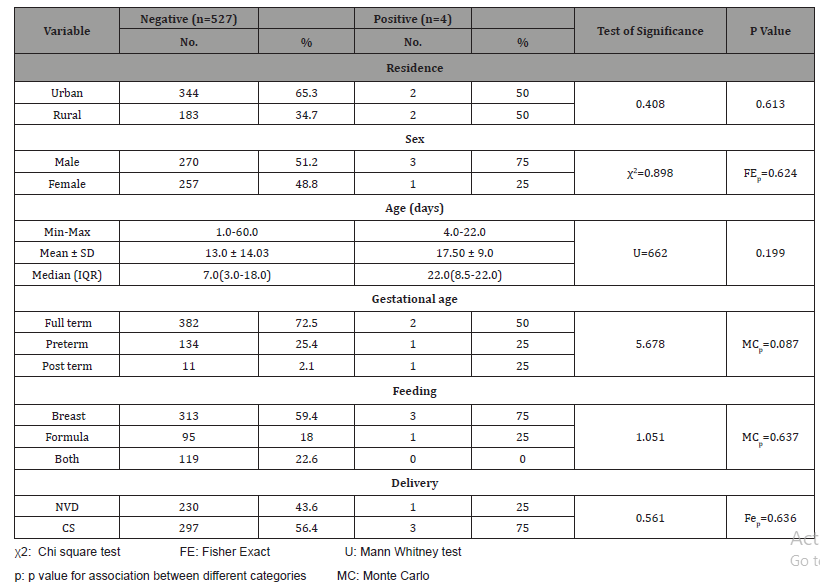
It reported that cases were collected over 14 months: there’s an increased rate of recruitment among the winter/fall season starting from (October-March) an average of 70 cases /month. While, summer months, the rate decreased down to 20 cases / months (Table 3).
This table shows the comparative study of the positive cases with S. Pneumonia carrier state to the negative group as regards the different demographic data. None of the studied variables was reported to be statically significant.
The height and weight of the studied population were not statistically related to the state of carriage of strept. Pneumonia.
Discussion
The colonization incidence differs from one country to another and changes before and after the introduction of PCV immunization in the same country.
In Egypt, the PCV vaccines were all licensed (PCV7-PCV10-and PCV13), but they were not incorporated in the NIP and vaccination is performed in the private medical clinics for those who can afford to pay for it.
Two randomized trials in Kenya and Papua New Guinea showed that PCVs are immunogenic and well tolerated when given at birth. However, the impact of neonatal or accelerated infant PCV schedules was noticed to be very limited on basis of vaccine serotype carriage and to low carriage in neonatal period. Pediatric nasopharyngeal flora is considered the largest reservoir for S. pneumoniae and nasopharyngeal carriage is a pre-requisite for infection.
It is clear that rapid uptake of any PCV disrupts the normal ecology of the colonized nasopharynx, and this impact both vaccine and non-vaccine serotypes organisms.
The best timing to start to give the vaccine might be beyond the first 60 days of age where the infant has very low carriage and has maternal antibodies protecting him from infection. Probably, vaccine given at this early age would be less effective in providing the required immunogenic response and fail to produce appropriate seroconversion. From all previously discussed literature review also we concluded that PCV13 would have a superior impact over the other vaccine types, causing protection against 13 serotypes included in it. And in addition, a protection against non-vaccine types through its higher cross-reactivity would be expected.
Conclusion
Low prevalence of carriage at 60 days of age and presence of more non- vaccine serotypes of pneumococci are two important conclusions reported in the present study.
Recommendations
From this study we recommend that: Wider study in different areas should cover all Egypt with a meta-analysis of the results from different areas, Further study in age group < 2 months healthy or have IPD.
Acknowledgement
None.
Conflict of Interest
No conflict of interest.
References
- Spijkerman J, Veenhoven RH, Wijmenga-Monsuur AJ, Elberse KE, van Gageldonk PG, et al. (2013) Immunogenicity of 13-valent pneumococcal conjugate vaccine administered according to 4 different primary immunization schedules in infants: a randomized clinical trial. JAMA 310(9): 930-937.
- Centers for Disease Control and Prevention (CDC) (2002) Recommended adult immunization schedule--United States, 2002-2003. MMWR Morb Mortal Wkly Rep 51(40): 904-908.
- Centers for Disease Control and Prevention (CDC) (2000) Preventing pneumococcal disease among infants and young children. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 49(RR-9): 1-35.
- Centers of Disease Control and Prevention (CDC) (2003) Drug-resistant Streptococcus pneumoniae disease. Atlanta, GA: CDC.
- Charan J, Biswas T (2013) How to calculate sample size for different study designs in medical research? Indian J Psychol Med 35(2): 121-126.
- Jonsson S, Musher DM, Chapman A, Goree A, Lawrence EC (1985) Phagocytosis and killing of common bacterial pathogens of the lung by human alveolar macrophages. J Infect Dis 152(1): 4-13.
- Bogaert D, Engelen MN, Timmers-Reker AJ, Elzenaar KP, Peerbooms PG, et al. (2001) Pneumococcal carriage in children in The Netherlands: a molecular epidemiological study. J Clin Microbiol 39(9): 3316-3320.
- Marchisio P, Claut L, Rognoni A, Esposito S, Passali D, et al. (2003) Differences in nasopharyngeal bacterial flora in children with nonsevere recurrent acute otitis media and chronic otitis media with effusion: implications for management. Pediatr Infect Dis J 22(3): 262-268.
- Norris CF, Mahannah SR, Smith-Whitley K, Ohene-Frempong K, McGowan KL (1996) Pneumococcal colonization in children with sickle cell disease. J Pediatr 129(6): 821-827.
- Coles CL, Kanungo R, Rahmathullah L, Thulasiraj RD, Katz J, et al. (2001) Pneumococcal nasopharyngeal colonization in young South Indian infants. Pediatr Infect Dis J 20(3): 289-295.
- Toltzis P, Jacobs MR (2005) The epidemiology of childhood pneumococcal disease in the United States in the era of conjugate vaccine use. Infect Dis Clin North Am 19(3): 629-645.
- Prymula R, Chlibek R, Splino M, Kaliskova E, Kohl I, et al. (2008) Safety of the 11-valent pneumococcal vaccine conjugated to non-typeable Haemophilus influenzae-derived protein D in the first 2 years of life and immunogenicity of the co-administered hexavalent diphtheria, tetanus, acellular pertussis, hepatitis B, inactivated polio virus, Haemophilus influenzae type b and control hepatitis A vaccines. Vaccine 26(35): 4563-4570.
- Russell FM, Biribo SS, Selvaraj G, Oppedisano F, Warren S, et al. (2006) As a bacterial culture medium, citrated sheep blood agar is a practical alternative to citrated human blood agar in laboratories of developing countries. J Clin Microbiol 44(9): 3346-3351.
- (2011) Laboratory Methods for the Diagnosis of Meningitis Caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae. WHO Manual.
-
Ashraf Khamis Mahmoud, Ahmed El Nawawy, Soad Farid Hafez, Manal Abdel Malek Antonios. Nasopharyngeal Colonization with Streptococcus Pneumoniae in Healthy Infants Less Than 60 Days Attending the Outpatient Clinic of Alexandria University Children’s Hospital. Glob J of Ped & Neonatol Car. 1(5): 2019. GJPNC.MS.ID.000522.
Streptococcus pneumoniae, Pneumococcal carriage, Healthy infants, Prevalence in less than 60 days, Pneumococcal serotypes, Penicillin sensitivity
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






