 Research Article
Research Article
Tar-Find Gene Extractor: A VIRmiRNA Tar-Find Companion Application for Extracting Gene Information
Daniela Chala Garcia1, Perpetua Muganda2, Clay Gloster3 and Christopher Doss4*
1Department of Biology, North Carolina Agricultural and Technical State University, USA
2Department of Biology, North Carolina Agricultural and Technical State University, USA
3Department of Computer Systems Technology, North Carolina Agricultural and Technical State University, USA
4Department of Electrical and Computer Engineering, North Carolina Agricultural and Technical State University, USA
Christopher Doss, Department of Electrical and Computer Engineering, 345 McNair Building North Carolina Agricultural and Technical State University Greensboro, USA.
Received Date: September 25, 2019; Published Date: September 27, 2019
Abstract
The VIRmiRNA Tar-Find web resource is available for the prediction of known and novel miRNA targets. Predicted miRNA targets are provided as hundreds to thousands of Ensembl transcript IDs, complicating and slowing the determination of the official gene symbol and function. We developed the Tar- Find Gene Extractor tool for the efficient, real-time conversion of VIRmiRNA Tar-Find miRNA target prediction results from Ensembl transcript IDs to official gene symbol and gene functions. The Tar-Find Gene Extractor operates through Microsoft ExcelTM, thus making this an easy to use tool for fast and efficient retrieval of Tar-Find predicted miRNA target genes’ information
Availability and implementation
The Tar-Find Gene Extractor Tool is available at Git Hub at https://github.com/ccdoss/tge.
Keywords: DAVID gene ID conversion tool; MiRNA target prediction; Ensembl database; Gene id retrieval; VIRmiRNA, Gene ID conversion
Abbreviations: TGE- Tar-Find Gene Extractor; Database for Annotation, Visualization and Integrated Discovery-DAVID
Introduction
VIRmiRNA Tar-Find is a user-friendly web tool dedicated to predicting target genes in human reference genes for both known and novel miRNA sequences [1]. VIRmiRNA Tar-Find is part of the VIRmiRNA web resource and is backed up by three databases comprised of 9133 experimentally validated records. The databases include experimental viral miRNAs, experimental miRNA targets, and experimental antiviral miRNAs [1]. VirmiRNA Tar-Find utilizes these databases to predict miRNA targets for known as well as novel miRNA sequences. Meanwhile, the web-based versions of , TargetScan [2], DIANA- TarBase v8 [3], PicTar [4], miRanda [5], etc., do not provide this novel miRNA target finding functionality [6,7].
VirmiRNA Tar-Find provides known and novel miRNA target prediction, and the identified miRNA gene targets are presented as hundreds to thousands of Ensembl gene transcript IDs. The format of the VIRmiRNA Tar-Find results complicates and slows down the determination of the official gene symbol and function. Manual gene ID conversion or the Database for Annotation, Visualization and Integrated Discovery (DAVID) gene ID conversion tool are candidates for official gene symbol determination. However, manual and DAVID Gene ID conversion methods have time and database availability limitations, respectively. Manual retrieval of gene names requires the user to access each VIRmiRNA Tar- Find provided hyperlink one by one to copy and paste the gene name into a new sheet. This process can take days or weeks, depending on the amount of target genes predicted, thus making it highly inefficient. DAVID (https://david.ncifcrf.gov/tools.jsp) is a web-based tool with different analysis capabilities, including conversion between multiple commonly used gene identifiers [8]. The DAVID Gene ID Conversion tool can perform the conversion of up to 3000 genes in a second, particularly when used outside of peak demand hours (The DAVID Knowledge Base. 2016; Huang da, 2008). However, when used during times of high demand, DAVID can produce no results or fail to complete the task. Moreover, DAVID is currently operating on the May 2016 release of Ensembl, even though the April 2019 release is currently available. Consequently, the DAVID ID conversion tool is currently limited by its out-of-date knowledgebase. Thus, we developed the Tar-Find Gene Extractor for the efficient, user-friendly retrieval of both official gene symbol and function in real time from Tar-Find miRNA target results as provided in form of Ensembl transcript IDs.
Materials and Methods
Development and features of the tar-find gene extractor
The Tar-Find Gene Extractor (TGE) is a companion application for the extraction of gene names and description information from VIRmiRNA Tar-Find results. TGE was developed by enabling developer mode within Microsoft Excel TM and utilizing Visual Basic for Applications capabilities. This facilitates the reading and writing of information within Microsoft ExcelTM sheets, as well as event driven activities, such as responding to changes in data or button clicks. The user enters the Tar-Find results into TGE, as shown in Supplemental Figure 1. The desired start and stop indices are then entered by the user. Possible values are 2 to N, where N is the total number of Tar-Find results (Row 1 is the header row, and therefore is not processed). After entering the indices, the user presses the Click Me button, which activates TGE. TGE operates in real time, accessing the Ensembl database and eliminating the need for any dependencies for operation. It is recommended users process between 500 and 1000 results at a time.
Testing Methodology
Ensembl transcript IDs predicted by VIRmiRNA Tar-find for hsa-mir21-5p miRNA were copied into a Microsoft Excel TM sheet. TGE was used to retrieve both official gene symbols and functions for the predicted transcript IDs. Percent of IDs converted (containing duplicate gene symbols), number of unique IDs converted (containing no duplicate gene symbols), convenience and accuracy of results were assessed. The results obtained by TGE were compared to results obtained using the DAVID ID conversion tool.
Results and Discussion
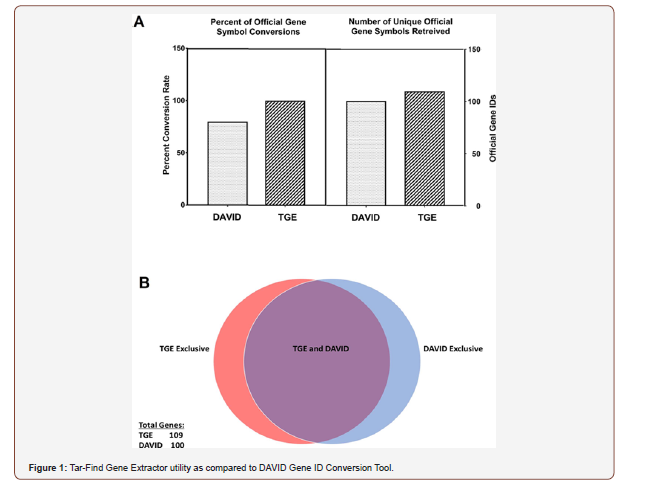
VIRmiRNA Tar-Find predicted 249 target genes with 100% match to nucleotides 1-10 of hsa- mir-21-5p miRNA; these were utilized for testing. TGE retrieved official gene symbol and description in an average of 6 minutes and 48 seconds. In contrast, the DAVID ID conversion tool failed to complete the conversion twice with an average waiting time of 7 minutes prior to manual program re-start. The DAVID ID conversion tool’s failure to produce results was likely due to the high traffic typically experienced by the platform. Following the second manual re-star of the DAVID tool, the ID conversion was completed in less than a minute TGE efficiently converted 249 gene IDs and provided their respective description. In comparison, DAVID only converted 198 IDs and provided no gene descriptions. Both TGE and DAVID converted IDs underwent sorting in Microsoft ExcelTM to remove duplicate gene IDs. A total of 109 unique gene IDs and descriptions were retrieved by TGE, and 100 unique gene IDs with no description were retrieved by DAVID (Figure 1A). In total, 97 unique gene IDs were identified by both TGE and DAVID, leaving an additional 12 gene IDs retrieved exclusively by TGE and 3 by the DAVID ID conversion tool (Figure 1B). The difference in functionality between TGE and DAVID are attributed to differences in the Ensembl database, where TGE utilizes the complete current release.
Conclusion
In summary, the Tar-Find Gene Extractor outperformed the DAVID gene ID conversion tool by retrieving 100% of input transcript IDs. The discrepancy noted between the unique IDs converted by TGE as compared to DAVID is likely explained by DAVID’s limited knowledgebase. While DAVID is operating on the May 2016 release of Ensembl, TGE accessed the Ensembl database in real time, and thus made use of the April 2019 release version. Finally, TGE provided the added benefit of generating brief Ensembl gene descriptions, while DAVID was limited to only performing ID conversions. Taken together, these results suggest TGE is a an efficient, convenient and reliable companion application for VIRmiRNA Tar-Find. (Supplemental Figure 1, Supplementary Data &Supplementary Data-Table).
Acknowledgement
The authors of this paper would like to acknowledge that this research has been supported by DoD grant W911NF1810445.
Conflict of Interest
The authors of this paper declare no conflict of interest.
References
- Qureshi A, Thakur N, Monga I, Thakur A, Kumar M (2014) VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets. Database (Oxford).
- Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4.
- Karagkouni, D, Paraskevopoulou MD, Chatzopoulos S, Vlachos I, Tastsoglou S, et al. (2018) DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 46(D1): D239-D245.
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, et al. (2005) Combinatorial microRNA target predictions. Nat Genet 37(5): 495-500.
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. (2004) Human MicroRNA targets. PLoS Biol 2(11): e363.
- Riffo Campos AL, I Riquelme, P Brebi Mieville (2016) Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int J Mol Sci 17(12).
- Oulas, A, Karathanasis N, Louloupi A, Pavlopoulos GA, Poirazi P, et al. (2015) Prediction of miRNA targets. Methods Mol Biol 1269: 207-229.
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. (2007) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35(Web Server issue): W169-75.
-
Christopher Doss, Daniela Chala Garcia, Perpetua Muganda, Clay Gloster. Tar-Find Gene Extractor: A VIRmiRNA Tar-Find Companion Application for Extracting Gene Information Glob J Eng Sci. 3(3): 2019. GJES.MS.ID.000564.
-
DAVID gene ID conversion tool, MiRNA target prediction, Ensembl database, Gene id retrieval, VIRmiRNA, Gene ID conversion, TGE- Tar-Find Gene Extractor, DAVID-Database for Annotation, Visualization and Integrated Discovery
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






