 Research Article
Research Article
Interactive Big Data Analytics Platform for Healthcare and Clinical Services
D Chrimes1*, MH Kuo2, AW Kushniruk2 and B Moa3
1Database Integration and Management, Vancouver Island Health Authority, Canada
2School of Health Information Science, University of Victoria, Canada
3Advanced Research Computing, University of Victoria, Canada
D Chrimes, Database Integration and Management, IMIT Quality Systems, Vancouver Island Health Authority, 1952 Bay Street, V8R1J8, Victoria BC Canada.
Received Date: August 20, 2018; Published Date: September 20, 2018
Abstract
A Big Data Platform (BDA) with Hadoop/MapReduce technologies distributed over HBase (key-value NoSQL database storage) and generate hospitalization metadata was established for testing functionality and performance. Performance tests retrieved results from simulated patient records with Apache tools in Hadoop’s ecosystem. At optimized iteration, Hadoop distributed file system (HDFS) ingestion with HBase exhibited sustained database integrity over hundreds of iterations; however, to complete its bulk loading via MapReduce to HBase required a month. The framework over generated HBase data files took a week and a month for one billion (10TB) and three billion (30TB), respectively. Apache Spark and Apache Drill showed high performance. However, inconsistencies of MapReduce limited the capacity to generate data. Hospital system based on a patient encounter-centric database was very difficult to establish because data profiles have complex relationships. Recommendations for key-value storage should be considered for healthcare when analyzing large volumes of data over simplified clinical event models.
Keywords: Adaptable architectures; Big data; Data mining; Distributed filing system; Distributed data structures; Healthcare informatics; Hospital systems; Metadata; Relational database
Introduction
Big Data is large, complex, distributed data collection growing extremely fast (or 5Vs- volume, variety, velocity, veracity, and value) [1,2]. At each and every attribute of Big Data there is high potential for unlocking new sources of economic values, providing fresh insights into scientific discoveries, and assisting on policy making [3]. However, Big Data is not practically useful until it can be aggregated and integrated into a manner that computer systems can generate knowledge. As data originally reside in individual silos, or across multiple domains that are not integrated, each source of knowledge serves a very specific localized purpose in the system or it is data entries for viewing only. Nevertheless, combination of data silos across data warehouses can provide new insights into important problems. Particularly, data correlation from multiple datasets can result in the identification of individual events and phenomena, as well as the creation of profiles to track all activities proactively.
Healthcare Big Data is different from other disciplines such as social network or business transitional data in that it includes structured Electronic Health Records (EHR) data, coded data, e.g., International Statistical Classification of Diseases (ICD) or Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT), semi-structured data (e.g. HL7 messages), unstructured clinical notes, medical images (e.g. MRI, X-rays), genetic and laboratory data, and other types of data (e.g. patient registration, public health and mental health data). Correspondingly, huge volumes and very heterogeneous raw data are generated by a variety of health information systems, such as electronic health record (EHR), Computerized Physician Order Entry (CPOE), Picture Archiving and Communication Systems (PACS), Clinical Decision Support Systems (CDSS), and Laboratory Information Systems (LIS). These system types are used in many distributed healthcare settings, such as hospitals, clinics, laboratories and physician offices. Several published studies have asserted if Big Data in healthcare is managed well, it can be used to drastically improve patient care services and reduce costs [3-6]. A McKinsey Global Institute study suggests, “If US healthcare were to use Big Data creatively and effectively to drive efficiency and quality, the sector could create more than $300 billion in value every year.” [7]. Shah & Tenenhaum [8] believe that Big Data-driven medicine will enable the discovery of new treatments for diseases. Garrison [9] also proclaims that Big Data will benefit population health and better decision making.
While the potential value of Big Data has been widely discussed, real-world applications and even application tests have rarely been done, especially those involving data from hospital systems. In our study, we designed and implemented a framework using Big Data technologies for supporting Big Data Analytics (BDA) in healthcare. As an applied platform, emulated patient data was distributed using Hadoop Distributed File System (HDFS) to simulate the interactive usability of extremely large volumes representing billions of patient encounters across multiple hospitals. To achieve this scale, three to nine billion patient records were constructed and cross-referenced with data profiles of metadata in existing data warehouses at the Vancouver Island Health Authority (VIHA), Victoria BC, Canada. It was then validated via workflow to retrieve results from patient queries. Overall the simulation was a proof-ofconcept implementation to use real patient data over BDA platform.
Literature Review
Challenges in big data analytics
Big Data Analytics (BDA) is the process of extracting knowledge or data mining from sets of Big Data [10]. Wang et al. [11] further described the extraction of useful knowledge from Big Data in terms of a processing pipeline that transfers, stores, and analyses data for whole systems. According to Kuo et al. [12], the process of achieving full Big Data utilization involves five distinct configuration stages; each stage has specific challenges, as follows:
Data aggregation
Copy/transfer data to a storage drive is the most commonly used method of aggregating and migrating large quantities of data but is not the most efficient with increasing volumes. Big Data usually involve multiple organizations, geographic locations, and multiple devices to aggregate over system; therefore, typically, generating large datasets from replication from production should minimize any ongoing consumption of network services and database resources affecting the production system to render the system in operation. Further, exchange of data between groups and databases is very difficult to coordinate; hence, a separate secondary database external from production systems for big data is usually carried out. Another approach to aggregate is to transfer data over a network. However, transferring, aggregating and indexing vast amounts of data require a significant bandwidth over a long duration. A third option is to replicate data from the sources and generate iteratively across instances and multiple nodes, as Hadoop does when replicating file blocks and storing them via distributed batch processes [13-15].
Data maintenance
Since Big Data involves large volumes, it is very difficult to store and maintain for ongoing queries, especially with continuous batch processes populating data. Moreover, time and cost can prohibit small organizations or departments from managing large amounts of data. Another challenge in healthcare is that real patient data, metadata, and data profiles need to constantly be updated with clinical events table; otherwise, the analytics is rendered useless. There are many solutions available to provide maintenance including cloud computing [16], grid computing [17], NoSQL/NewSQL and other storage systems (e.g., MongoDB, HBase, Voldemort DB, Cassandra, Hadoop Distributed File System (HDFS) and Google’s Bigtable [18-20]).
Legality and ethics are a major issue in data maintenance. Security, confidentiality and privacy mandated by legislation and strict regulations are key data governance that holds maintenance accountable. For example, the Health Insurance Portability and Accountability Act require the removal of 18 types of identifiers, including any residual information that could identify patients. Privacy concerns can be addressed by using software’s and database technologies, such as key-value storage services, but advanced configuration and technical knowledge is needed. For example, Pattuk et al. [21] proposed a framework for secure Big Data management involving an HBase database called Big Secret securely outsourced and processed encrypted data over public keyvalue stores. Derbeko et al. [22] also provided a comprehensive review of existing security and privacy protocols for distributed processing of large-scale data on a Cloud computing environment. Although most hospitals securely house their data in server racks and the vendors commonly are not allowed to use cloud services, especially when there is no control of the location and if maintenance costs increase.
Data integration
Data integration and interoperability processes involve combining (possibly transforming) data into an appropriate format for analysis. Since Big Data in healthcare are extremely large, distributed at different locations, unstructured and heterogeneous, the management of data integration over time is very challenging [16,23]. Numerous solutions have been proposed for raw Big Data integration [24-28]. These methods are problem-oriented, i.e., the method is only applied to specific data sets or aggregates. Very few generic approaches exist for integrated unstructured data.
Data analysis
BDA complexity involves analytic algorithms to be programmed. However, with increasing complexity, computing time increases dramatically even with small increases in data volume. For example, Bayesian Networks are a popular algorithm for modeling knowledge in computational biology and bioinformatics, and the computing time required to find the best network increases exponentially as the number of records rises [29]. Even for simple data analysis, it can take several days; even months, to obtain a result when databases are very large and SQL-like “joins” are executed. Moreover, many studies suggest parallelization of computing model for high performance over platforms with reduced computationally intense problems during analysis [30-37].
Pattern interpretation
Knowledge representation is an absolute necessity to achieve for any data mining and BDA platforms in healthcare. Further, BDA is of little value if decision-makers do not understand the patterns to find a discovery. Additionally, given the complex nature of Big Data, representations of trends and individualized results will not be comprehensible to non-experts initially using the platform. Moreover, many people instinctively believe that bigger data means better information. Additionally, agile data science can offer accurate data visualizations but cannot protect us from inaccuracies and faulty assumptions. Many reporters are often fooled into thinking that correlations have true significance (or lack thereof) without hidden nuances in the data, its quality, and its structure.
Big data technologies and platform services
Big Data technologies fall into four main categories: highperformance computing (HPC), storage, resource/workflow allocator, and insertion or ingestion processes [36]. An HPC system is usually the backbone to the technology framework (e.g., IBM’s Watson). HPC can consist of a distributed system, grid computing, and a graphical processing unit (GPU). In a distributed system, several computers (computing nodes) can participate in processing large volumes and variety of structured, semi-structured, and/or unstructured data. A grid computing system is a distributed system employing resources over multiple locations (e.g., CPUs, storage of computer systems across network, etc.), which enables processes and configurations to be applied to any task in a flexible, continuous, and inexpensive manner. GPU computing is well-adapted for throughput-oriented workload problems, like batch fills at large volume. Parallel data processing can be handled by GPU clusters. However, “GPUs have difficulty communicating over a network, cannot handle virtualization of resources, and using a cluster of GPUs to implement a commonly used programming model (namely, MapReduce) presents some challenges” [36].
In the quest for the most effective and efficient platform, distributed systems appear to be the best choice of new technologies in the near future. Therefore, it is important to understand the nature of a distributed system, as compared to conventional grid computing, which can also be applied to supercomputing and high-performance computing, and this does not necessarily mean data mining or big data. Furthermore, a distributed computing system can manage hundreds to thousands of computer systems, each of which is limited in its processing resources (e.g., memory, CPU, storage, etc.). By contrast, a grid computing system makes efficient use of heterogeneous systems with optimal workload management servers, networks, storage, and so forth. Therefore, a grid computing system supports computation across a variety of administrative domains, unlike a traditional distributed system.
The most common computing uses a single processor (in desktop personal computers), with its main memory, cache, and local disk (or a computing node). In the past, applications used for parallel processing for large scientific statistical calculations employed special-purpose parallel computers with many processors and specialized hardware. However, the prevalence of large-scale web services has encouraged a turn toward distributed computing systems: that is, installations that employ thousands of computing nodes operating more or less independently with an application (e.g., [38,39]). These computing nodes are off-the-shelf hardware, which greatly reduces the cost of distributed systems (compared to special-purpose parallel machines) and create localized interactive access to BDA platforms. Thus, meeting the needs of distributed computing, a new generation of programming frameworks has emerged. These frameworks take advantage of the power of parallelism while avoiding pitfalls, such as the reliability problems posed by the use of hardware consisting of thousands of independent components, any of which can fail at any time. In our framework, the Hadoop cluster, with its distributed computing nodes and connecting Ethernet switch, runs jobs controlled by a master node (known as the Name Node), which is responsible for chunking data, cloning it, sending it to the distributed computing nodes (known as Data Nodes), monitoring the cluster status, and collecting or aggregating the results [38]. It, therefore, becomes apparent that not only the type of architecture and computing system is important for a BDA platform but also the inherent versus customizable processes in the data processes to produce results.
A number of high-level programming frameworks have been developed for use with HDFS; MapReduce, a programming framework for data-intensive applications proposed by Google, is the most popular component with Hadoop and its utilization has increased. Borrowing from functional programming, MapReduce enables programmers to define Map and Reduce tasks to schedule process large sets of distributed data in specific sequence or modified output, thus allowing many of the most common calculations on large-scale data to be performed on computing clusters efficiently and in a way that is tolerant (even fault tolerant) of hardware failures during compaction/computation. Therefore, distributed computing using MapReduce and Hadoop framework represents a significant advance in the processing and utilization of Big Data, especially in banking and transportation sectors and even in healthcare [36,40]. However, MapReduce is not suitable for online transactions or streaming, and, therefore, the system architecture may not be compatible if real-time data is required to retrieve accurately.
MapReduce programming framework has high degree of parallelism combined with its simplicity and its applicability to a wide range of domains. The degree of parallelism depends on the size of the input data [20]. The Map function processes the inputs (e.g., key1, value1), returning other intermediary pairs (e.g., key2, value2). Then the intermediary pairs are grouped together according to their keys. The Reduce function outputs new key-value pairs (e.g., key3, value3). High performance is achieved by dividing the processing into small tasks run in parallel in a cluster. Programs written in a functional style are automatically parallelized and executed by MapReduce. Employing MapReduce and Hadoop has two advantages: (1) reliable data processing via fault-tolerant storage methods that replicate computing tasks and clone data chunks on different computing nodes across the computing cluster, and (2) high-throughput data processing via a batch processing framework and the Hadoop Distributed File System (HDFS) [38,40- 43]. Thus, it is apparent that not only is Hadoop and MapReduce compatible but also their inherent processes are important for the platform to perform well.
A large and growing range of technologies related to Hadoop are emerging. Open-source software for use with the HDFS is constantly being updated monthly and even weekly. Chang et al. [41] stated that HDFS is designed to deal with data for building a distributed data center mostly from software not hardware. For instance, software installed for versions of HDFS uses data duplicates to enhance reliability. However, data duplicates need extra storage space and hence a larger investment in infrastructure. On the other hand, deduplication techniques can improve efficiency in the use of storage space [41]. Even with the number of software versions in Hadoop’s ecosystem increasing, there is more software and big data platforms applied to bioinformatics and genetics than hospital systems. Very few big data platforms are applied to realtime utilization in hospitals globally. Relatively new software like Apache Spark (est. 2013) and Apache Drill (est. 2015) has not been tested for clinical databases and systems. Furthermore, very little is known about the usability of big data platforms or frameworks in healthcare.
Design the Analytics Framework
In considering the design of a framework for a BDA platform to use in the field of healthcare, the basic construct of processes over a platform requires diverse clinical data from multiple sources and querying large volumes. Also, the configuration must ensure patient data security, confidentiality, and privacy. This section describes our approach of constructing the framework.
The conceptual analytics framework
The BDA platform should harness the power of accessible front-end applications to analyze large quantities of data in an interactive manner, while enriching the user experience with data visualizations. All this functionality must be accomplished at moderate expense. Based on these requirements, we construct an interactive dynamic framework (Figure 1). And to meet requirements of clinical users front-end and interfaced applications were tested (i.e. Apache Phoenix, Spark, and Drill) and integrated with the HDFS and back-end NoSQL database of HBase. Together, the framework and the applications allow users to query, visualize, interpret, and modify outputs of the data. The overall purpose is to make Big Data capabilities accessible to stakeholders, including University of Victoria (UVic) researchers, VIHA physicians, healthcare practitioners, database administrators, and webbased application developers. The proposed analytics framework included the following nine components:

• A high-performance computer (HPC) clusters with a total of 72 cores and parallelized six nodes. Serial programs run on one CPU or core in one of the computing nodes in the cluster. Multiple copies are run over the cluster by submitting serial batch jobs by user; while parallel programs, on the other hand, had multiple processes or threads that can run at the same time. This study utilized the batch serial process to access and start jobs over the Hadoop with some parallel MapReduce processes in the top-down architecture from head node to worker or slave nodes.
• A back-end NoSQL database of HBase (NoSQL version 0.98.11) was used. HBase is a NoSQL database composed of the main deployment master (DM) node and its fail-over master, the Region Servers holding HBase data, and Zookeeper, which contained services to allocate data locality [44], of three nodes, that orchestrated that ensemble. The xml configuration files of HBase-site.xml and the HBase-env.sh were adjusted to improve the performance of HBase. HBase was chosen due to its NoSQL services, especially linear to modular scalability to document architecture. It uniquely indexed rows to columns and established key-stores to encrypt the data. In addition, it allowed for SQL-like layer of Apache Phoenix to be configured on top of HBase big-tables.
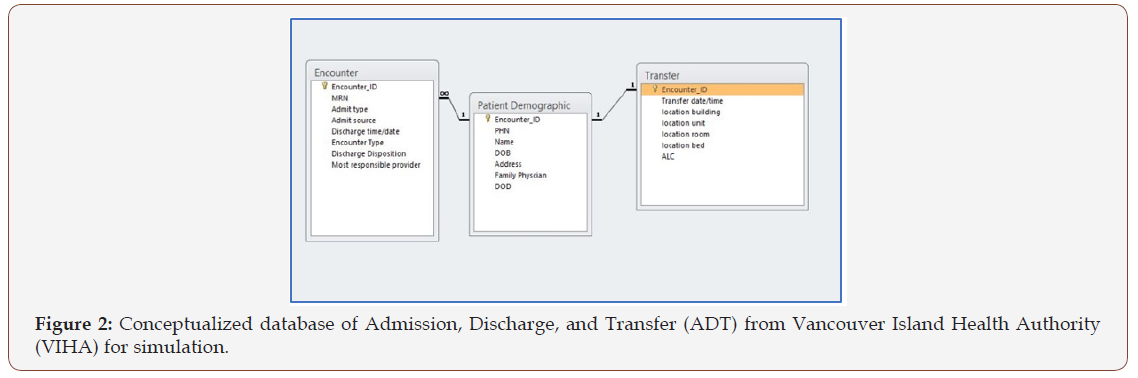
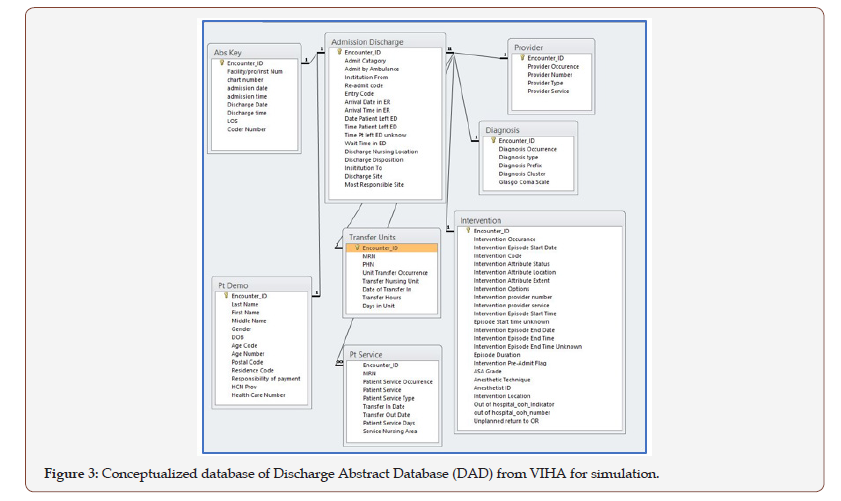
• In the emulation of the database, each row represented encounter-based patient data as a Big Integer type with diagnoses, interventions and procedures specific to that patient. This patient encounter model coincided with the Health Authority’s current Admission, Discharge, and Transfer (ADT) system in its database schema linked to data warehouses, which also includes Discharge Abstract Database (DAD) (see Table 1). This patient-specific structure in the database allows for active updates to add to the data generation while maintaining accurate patient querying over the platform in the system. All necessary data fields were populated for 50 million records before replication to form three-nine billions of records. This data followed the workflow of VIHA staff. Rows across the columns according to the existing abstraction of patient visits to hospital. HBase established this row-column relationship(s) with a wide range of indexes for each unique row, and each row contained a key value that was linked to the family of qualifiers and primary keys (columns). HBase operators were specific to family qualifiers at each iteration; therefore, the data was patient-centric combined with certain data (from different sources of metadata), such that summary of diagnosis or medical services as a whole could be queried.
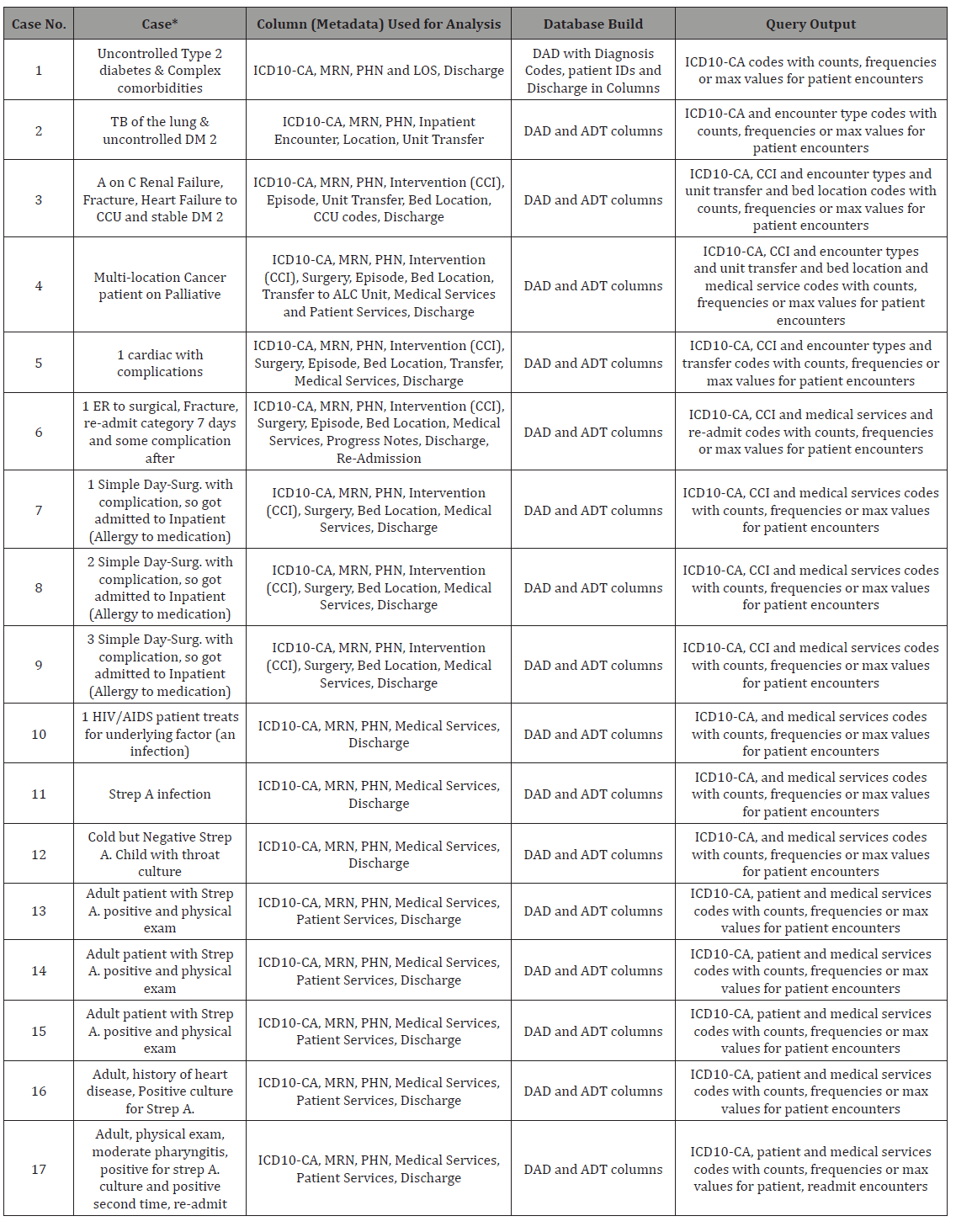
Table 1: Use cases and patient encounter scenarios related to metadata of the patient visit and its placement in the database related to query output.
• The construction of the framework of HBase (NoSQL) across Hadoop (HDFS version 2.6.0) \MapReduce established the BDA platform. This construct coincided with and was enforced by the existing architecture of the WestGrid clusters at UVic (secure login via lightweight directory access protocol or LDAP to deploy jobs under restricted accounts). It was initially running the architecture of the platform with five worker nodes and one master node (each with 12 cores) and planned on increasing the (dedicated) nodes to 11 and possibly to 101, as well as incorporating a non-dedicated set of virtual machines on WestGrid’s open stack cloud.
• A master deployment manager (DM) is the portable batch serial login that was configured as head node to worker nodes. Deployment of the Hadoop environment on the nodes carried out via a sequence of setup scripts that the user calls after loading the necessary modules and setup additional configuration to the head node with YARN and Zookeeper as allocators of various deployments. Setup scripts created an initial configuration depending on the number of nodes chosen when launching the job. The user can adjust those configurations to match the needs of the job and its performance. The data were distributed in parallel on the nodes via a balanced allocation with every running part of the batch jobs in an initialized serial computing process.
• Making the DBA platform InfiniBand-enabled was challenging, as most of the Hadoop environment services rely on the hostname to get the IP address of the machine. Since the hostnames on a cluster are usually assigned to their management network, the setup scripts and the configuration files required adjustment. The InfiniBand was used because it offers low latency and high bandwidth (~40Gb/s) connectivity between the nodes. YARN, Hadoop’s resource job manager, unfortunately still partly uses the Gig-Ethernet interface when orchestrating processes among the nodes, but the data transfer was carried out on the InfiniBand. Yarn was the resource manager of Hadoop and services of scheduling incongruent to running the Hadoop jobs [38].
• The queries via Apache Phoenix (version 4.3.0) resided as a thin SQL-like layer on HBase. This allowed ingested data to form structured schema-based data in the NoSQL database. Phoenix can run SQL-like queries against the HBase data. Similar to the HBase shell, Phoenix is equipped with a python interface to run SQL statements and it utilizes a .csv file bulk loader tool to ingest a large flat file using MapReduce. The load balancing between the Region Servers (e.g., “salt bucket” code) was set to the number of worker nodes that allowed ingested data to be distributed evenly.
• Apache Spark (version 1.3.0) was also built from source and installed to use on HBase and the Hadoop cluster. Spark utilizes Yarn and HDFS architecture and is known to scale and perform well in the data space (over distributed files).
• Inspired by Google’s big query engine Dremel, Drill offers a distributed execution environment for large-scale ANSISQL: 2003 queries. It supports a wide range of data sources, including .csv, JSON, HBase, etc... [45]. By (re)compiling and optimizing each of the queries while it interacted with the distributed data sets via the so-called Drill bit service, Drill showed capacity of the query with performance at a low latency SQL query. Unlike the master/slave architecture of Spark, in which a driver handles the execution of the DAG on a given set of executors, the Drill bits were loosely coupled and each could accept a query from the client [46]. The receiving Drill bit becomes the driver for the query, parsing, and optimization over an efficient, distributed, and multiphase execution plan; it also gathers the results back when the scheduled execution is done [46, 47]. To run Drill over a distributed mode, the user will need a Zookeeper cluster continuously running. Drill 1.3.0 and Zookeeper 3.4.6 were installed and configured on the framework of the platform over a port with a local host.
Computational platform
In this study, we used high performance clusters of WestGrid. WestGrid is a Canada government-funded infrastructure program started July 2010 at UVic. In Western Canada, it provides access to high performance computing and distributed data storage, using a combination of grid, networking, and collaboration tools [48].
The WestGrid computing facilities at UVic house the Hermes and Nestor system, which are the two super clusters that share infrastructure, such as resource management, job scheduling, networked storage, and service and interactive nodes. Also, Hermes and Nestor share login nodes: hermes.westgrid.ca and nestor.westgrid.ca (hermes.westgrid.ca and nestor.westgrid.ca are aliases for a common pool of head nodes named litaiNN.westgrid. ca). Nestor is a 2304-core capability cluster geared towards parallel jobs. It consists of 288 IBM iDataplex servers with eight 2.67 GHz Xeon x5550 cores and 24 GB of RAM. Data is shared between nodes and the GPFS file system using a high-speed InfiniBand interconnect. Hermes is a 2112-core capacity cluster geared towards serial jobs. It consists of 84 nodes having eight cores each and 120 nodes with 12 cores each.
The data process pipeline
The proposed framework with the techniques and tools used in a data process pipeline were carried in six steps.
Step 1: Data preparation and privacy protection: Privacy protection is an important requirement in healthcare data. All patient data must be identifiable and cataloged across the hospital system. Typically, this is carried out by business/security analysts based on regulations and data analyst for maintenance in data warehouse. The goal of this step (in our study) is to generate a comprehensive list of sensitive elements specific to the organization and discover the associated tables, columns and relationships across the data warehouse. After that is established, data masking or generating an encrypted data replication over its distribution will be used to conceal the patient’s identity while preserving the information, relationships, and context [49].
Step 2: Data acquisition: There is a wide range of tools and technologies for bulk loading and accessing large datasets from different sources. In this study, we proposed to use Apache Sqoop to ease the transfer between VIHA data warehouse and the WestGrid. Sqoop is a tool designed for efficiently transferring bulk data between Apache Hadoop and structured data repositories. For collecting and gathering unstructured data, such as logs, we used Apache Flume. Flume is a distributed, reliable, and available service for efficiently collecting, aggregating, and moving large amounts of log data. High-speed file transfer technologies, such as SCP and GridFTP, were used to transfer huge amounts of data into the parallel file system (GPFS).
Step 3: Data maintenance: The aggregated data was stored and maintained over HDFS in NoSQL HBase over Hermes nodes in WestGrid. To improve the ingestion of the 90 columns over generated three-nine billion rows, local disks of 40TB in total were physically installed on the worker nodes. After local disks were installed, a set of shell scripts was used to automate the generation and ingestion process of 50 million records as a batch at each of the iterations (via MapReduce). The goal of study was to run towards ten billion rows but due to operational barriers, workflow limitations and table space of key stores, which almost tripled during the iterations, nine billion achieved but only three billion could be queried. Some limitations of the Hadoop/MapReduce framework occurred because local disks had to be reinstalled after the failover from WestGrid’s 500GB disks to 2TB slots did not work.
Step 4: Data integration: The data were modelled according to the critical data profiles from the clinical event tables. Each of the data elements were derived from naming conventions of the patient record tables of Admission, Discharge, and Transfer (ADT), i.e., Figure 2, as well as data standard of Discharge Abstract Database (DAD), i.e., Figure 3, for clinical reporting. This was technically established by SQL-like code run over HBase via Apache Phoenix [50]. Additionally, the most important step in the data integration was that the data was transformed into appropriate formats via task scheduler in MapReduce over HDFS. The two components Map and Reduce placed files over the distributing file process but there were different performances. Map portion took only three minutes, but the Reduce part of bulk load and indexing took three to twelve hours. Also, reduce did not run at a constant percentage of Map components and ranged from 20-60%. To improve the Reducer, several manual steps were taken including compression of the HFiles in HBase. However, there was still on average 400-500 minutes to complete the upload and integration, and the compression failed periodically, which resulted in a delay to clean out failed files and re-process the data from MapReduce. This issue was investigated and deemed caused in large part by HBase’s Region Server hot-spotting or over-utilization that stalled the releasing process. Thus, the data process of the platform had a major limitation with the MapReduce that persisted. Additionally, as part of the process, Zookeeper and Yarn connectivity and Region Servers connecting to network only persistently connected to UVic’s network instead of WestGrid’s InfiniBand, which drastically slowed performance. Some configuration to Zookeeper and Yarn minimized this occurrence but no concrete resolution was found. Thus, this step to integrate the data over the HDFS to HBase required manually configuration and monitoring at each and every batch submitted.
Step 5: Data analysis: To handle intensive computation, we used the MapReduce programming model that distributed analytic/ data mining tasks. The MapReduce templates were tailored to use with applications of Apache Phoenix, Spark and Drill on the HBase database. To give the structured data the same semantics, as its relational database counterpart, we opted to use Phoenix on HBase because it is still a relational database layer that runs on top of HBase and offers a low-latency SQL-like access to HBase by running multiple parallel HBase scans on different region servers [14]. For developing some of the applications for end users, we also engineered notebooks on Jupyter and Zeppelin interfaces over Web browsers to connect to the database and interactively apply queries simultaneously to generate results.
Step 6: Pattern representation and visualization: Pattern presentation was found thru interface tools of Apache Spark (version 1.3.0) and Drill over the platform. Apache Spark was also built from source and installed to use on HBase and the Hadoop cluster. The intent was to compare different query tools like Apache Spark and Drill against Apache Phoenix using similar SQL-like queries. The results showed that the ingestion time of one billion records took circa two hours via Apache Spark [51]. Apache Drill outperformed Spark/Zeppelin and Spark/Jupyter. However, Drill was restricted to running more simplified queries, and was very limited in its visualizations. Therefore, it exhibited poor usability for healthcare. Zeppelin, running on Spark, showed ease-of-use interactions for health applications, but it lacked the flexibility of its interface tools and required extra setup time with a 30-minute delay before running queries. Jupyter on Spark offered high performance stacks not only over our platform but also in unison to run all queries simultaneously for a variety of reporting for providers and health professionals.
System Simulation
Computing platform and configurations
In this section, we describe our steps and experiences to test the technical framework of the BDA platform. To accomplish this, we installed and configured a Hadoop environment from source on the WestGrid cluster, and a dynamic Hadoop job was launched. It was configured by hdfs-site.xml and a MapReduce frame, configured via MapRed-site.xml that ran under the Hadoop resource manager Yarn (with configuration file yarn-site.xml). The number of replicas was set to three. To interact with HDFS, command scripts were run to automate the ingestion step (generate data replication in the exact format specified by SQL script to the nodes).
The BDA platform was built on top of the available open source software called HBase. HBase comprised a main deployment master (DM) and fail-over master. Its region servers held HBase data, and a Zookeeper of three nodes orchestrated the ensemble. The xml configuration file HBase-site.xml and the HBase-env.sh were adjusted to configure and fine tune HBase. HBase was chosen due to its real-time read/write access, and linear and modular scalability. HBase consisted of unique rows and each row contains a key value. A key value entry has five parts: row-key (row), family (fam), qualifier (qua), timestamp (ts) and value (val) denoted as:
KEY: = row||fam||qua||ts.
Additionally, to establish the HBase key value entries there are four operations:
• Put – inserts data;
• Get – retrieves data of a specific row;
• Delete – removes a data row;
• Scan – retrieves a range of indexed rows.
The last three operations can be limited to specific family, qualifiers, or over a certain time range. In addition, it allows for SQL-like layers via Apache Phoenix to be configured on top to bulk load to HBase [50]:
CREATE TABLE IF NOT EXISTS DADS1 (
EncounterID BIGINT NOT NULL,
Admit_by_Ambulance VARCHAR,
Admit_Category VARCHAR,
Admission_Date VARCHAR,
Admission_Time VARCHAR,
age INTEGER,
Anesthestic_Grade VARCHAR ,
Anesthetist_ID VARCHAR,
Anesthetistic_Technique INTEGER,
Arrival_Date_in_ER VARCHAR NOT NULL,
Arrival_Time_in_ER VARCHAR NOT NULL,
Date_Patient_Left_ED VARCHAR ,
Date_of_Transfer_In VARCHAR,
Days_in_Unit VARCHAR,
Discharge_Date VARCHAR NOT NULL,
Discharge_Disposition VARCHAR NOT NULL,
Discharge_Site VARCHAR NOT NULL,
Discharge_Time VARCHAR NOT NULL,
Birth_Date VARCHAR,
Diagnosis_Cluster VARCHAR,
Diagnosis_Code VARCHAR NOT NULL,
Diagnosis_Occurrence INTEGER,
Diagnosis_Prefix VARCHAR,
Diagnosis_Type VARCHAR,
Entry_Code VARCHAR,
Episode_Duration INTEGER,
First_Name VARCHAR,
Glasgo_Coma_Scale VARCHAR,
Gender VARCHAR,
Health_Care_Number VARCHAR NOT NULL,
HCN_Prov VARCHAR,
Institute_From INTEGER,
Institution_To INTEGER,
Interven_Attribute_Extent VARCHAR,
Interven_Attribute_Location VARCHAR,
Interven_Attribute_Status VARCHAR,
Interven_Code VARCHAR,
Interven_Episode_Start_Date VARCHAR,
Interven_Episode_St_Date VARCHAR,
Interven_Location VARCHAR,Interven_Occurrence INTEGER,
Interven_Options VARCHAR,
Interven_Pr_Number INTEGER,
Interven_Preadmit_Flag VARCHAR,
Interven_provider_service INTEGER,
Interven_Start_Time_unknown VARCHAR,
Last_Name VARCHAR,
LOS INTEGER NOT NULL,
Middle_Name VARCHAR,
Most_Responsible_Site VARCHAR,
MRN VARCHAR,
Out_of_Hospital_ooh_indicator VARCHAR,
Out_of_hospital_ooh_number INTEGER,
PHN INTEGER,
Postal_Code VARCHAR,
Pdr_Number INTEGER,
Provider_Occurrence INTEGER,
Provider_Service VARCHAR,
Provider_Type VARCHAR,
Patient_Service INTEGER,
Patient_Service_Days INTEGER,
Patient_Service_Occurrence INTEGER,
Patient_Service_Type VARCHAR,
Readmit_Code INTEGER,
Reporting_Prov INTEGER,
Residence_Code INTEGER,
Responsibility_for_payment INTEGER,
Service_Nursing_Area VARCHAR,
Time_Patient_Left_ED VARCHAR,
Time_Pt_left_ED_unknown VARCHAR,
Transfer_Hours VARCHAR,
Transfer_Nursing_Unit VARCHAR,
Transfer_In_Date VARCHAR,
Transfer_Out_Date VARCHAR,
Unit_Transfer_Occurrence INTEGER,
Unplanned_return_to_OR VARCHAR,
Wait_Time_in_ED VARCHAR,
FIN INTEGER NOT NULL,
Encounter_Number INTEGER NOT NULL,
Admit_Source VARCHAR,
Encounter_Type VARCHAR,
Medical_Services VARCHAR,
MostResponProvider VARCHAR,
Address VARCHAR,
Family_Physician VARCHAR,
Location_Building VARCHAR NOT NULL,
Location_unit VARCHAR NOT NULL,
Location_Room VARCHAR NOT NULL,
Location_Bed VARCHAR NOT NULL,
CONSTRAINT PK PRIMARY KEY (EncounterID, Arrival_Date_ in_ER, Arrival_Time_in_ER, Discharge_Date, Discharge_Disposition, Discharge_Site, Discharge_Time, Diagnosis_Code, Health_Care_ Number, OS, FIN, Encounter_Number, Location_unit), Salt Buckets =5.
The deployment of the Hadoop environment was carried out via a sequence of setup scripts that the user runs after loading modules. These setup scripts created an initial configuration over the number of nodes requested to the batch system. The user could adjust the configurations to match the needs of the job. The services that the master node and slave nodes run are shown in Figure 4.
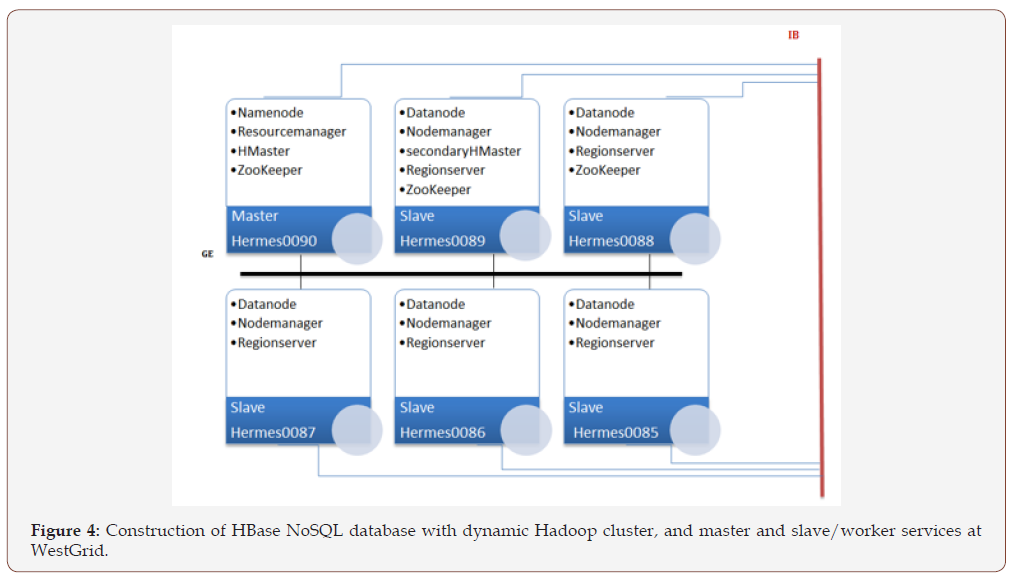
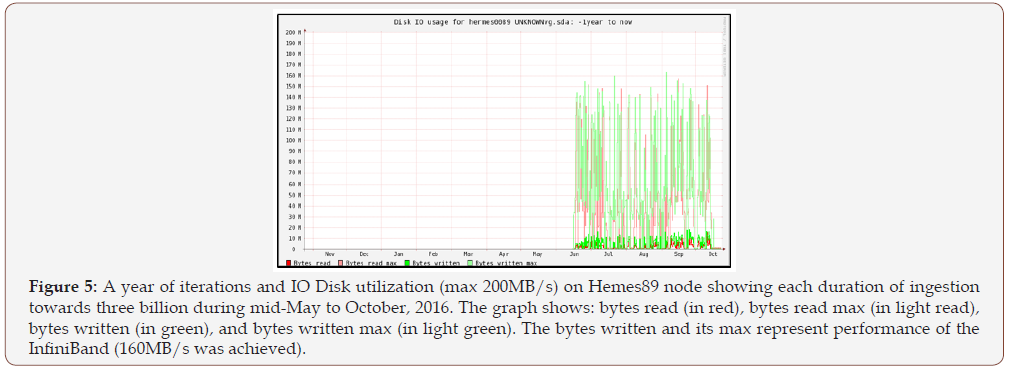
Making the platform InfiniBand-enabled was challenging. The InfiniBand was desirable as it offered low-latency and highbandwidth (~40Gbit/s) connectivity between the nodes. However, most of the Hadoop environment services rely on the hostname to get the IP address of the machine. Since the hostnames on a cluster are usually assigned to their management network, the setup scripts and the configuration files needed to be adjusted. Yarn, Hadoop’s resource and job manager, unfortunately, still uses the Gig-Ethernet interface when orchestrating between the nodes but the data transfer is still carried out over the InfiniBand. Figure 5 shows peak values in disk space on a Hermes node utilized that is indicative of the maximum reached at the start of file ingestion via InfiniBand. The lows are due to compaction running on the disks after disk space is maximized and this was necessary to maintain a consistent range of data distributed.
The Apache Phoenix allowed the ingestion to set a structured schema into the NoSQL database. The usual SQL queries can run in Phoenix against the HBase data. Similar to HBase shell, Phoenix is equipped with a python interface to run SQL statements. Moreover, Phoenix comes with a Comma-Separated Value (CSV) file bulk loader tool to ingest the large flat CSV files using MapReduce into a big table. Due to throughput and limited wall time, we could not ingest the full 1 billion records using HBase in one run (using the bulk loader) as its MapReduce required more than the available local disk spaces on the nodes to store the intermediate temporary files and timed out. Batch jobs (twenty-eighty) of 50-million records were then ingested one at a time. As explained further in the results, even with that, many ingestion attempts took a longer time than expected or failed. The load balancing between the Region Servers was also a major issue. Additionally, the performance and partitioning of data by MapReduce (its mapping and sorting) is highly dependent on how evenly it can distribute the workload [19,50]. To address it, we had to pre-split the table using the Phoenix “salt bucket” mechanism to evenly balance the distributed data to the nodes.
Data emulation and modeling
The established BDA platform was used to benchmark the performance of mining current and future reporting of VIHA’s clinical data warehouse, which in archive spans more than 50 years. Currently, the data warehouse has 100 facts and 581-dimension tables that all encompass a total of 10 billion records. Huge volumes of health data are continuously generated and added to this collection. Within the data warehouse, two of the largest datasets are the ADT and DAD. The former contains individual patient bedtracking information, while the latter contains Canadian Institute for Health Information (CIHI) diagnostic codes and discharge abstract metadata fields.
Over the span of twelve months in 2014-15, several interviews were conducted with business intelligence data warehouse, clinical reporting, database administrators, application platform, and health informatics architecture teams employed at VIHA. All interviews were recorded and documented. During these interviews, a patient data system generated from hospital admissions (based on encounter types) and a discharge system (based on diagnoses and procedures) were established. Furthermore, data profiles, including dependencies and the importance of the metadata for the reporting performance of the hospital, were confirmed and verified for use over the Hadoop/MapReduce to HBase framework.
In the emulated datasets, each row represented encounterbased patient data, with diagnoses, interventions, and procedures specific to that patient, that the current ADT system has in its database schema linked to a bigger data warehouse, which includes DAD (refer to Table 1). This patient-specific structure in the database allowed for active updates for accurate querying. The data emulated for both ADT and DAD were greatly simplified from the real patient records of hospitals (refer to Figures 2 & 3). In fact, the hospitals ADT alone has over 100 tables that could have patient information to query. However, we cross-referenced the most important information from ADT and DAD for clinical reporting with data wanting to be queried as a combined large data.
Unlike relational database systems, HBase itself does not support SQL; in fact, HBase is not a relational data store at all. HBase applications like Apache Phoenix can provide an SQL-like layer over HBase and are written in Java, much like typical MapReduce applications. HBase settings had to be purged or cleaned after each of the ingestions due to unknown tracers or remnants of transactions that then later caused query inaccuracies. Every HBase system components comprises a set of tables. Each table must have an element defined as a Primary Key, and all access attempts to HBase tables need to use this Primary Key; even Apache Spark SQL can be linked to database conversions with HBase [52], and other transformation methods can be applied to healthcare data towards an intact NoSQL database [53]. Additionally, in our study, HBase column represented an attribute of an object; for example, if the table had logs from servers, where each row might be a log record, a typical column would show the timestamp of the time when the log record was written, or perhaps name of the server on which the record was originated. Thus, HBase allowed for many attributes to be grouped into so-called column families and stores the elements of the column family together.
Performance evaluation
The pathway to running ingestions and queries from our build of the BDA platform was as follows: CSV flat files generated → HDFS ingestion(s) → Phoenix bulk loads into HBase →Apache Phoenix Queries. Under this sequence and after loading the necessary module environments for Hadoop, HBase and Phoenix and testing initial results linked to the family qualifiers and HBase key value entries. The SQL code (shown in Table 1) was then iteratively run to ingest 50 million rows to the existing NoSQL HBase database (i.e., names of the columns were shortened but metadata kept exactly the same as ADT and DAD tables shown in Figures 2 & 3).
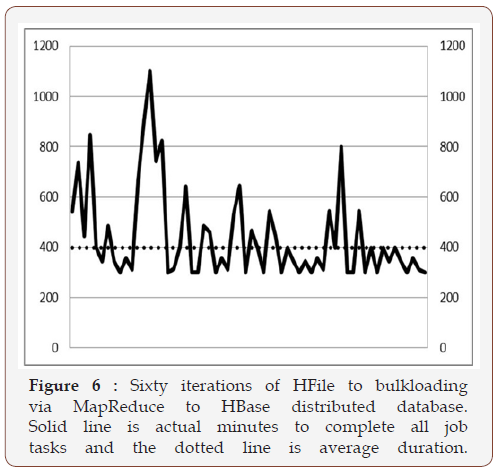
For performance benchmarking, three metric measures were used: HDFS ingestion(s), bulk loads to HBase, and query times via Phoenix. Three flat files in CSV format were ingested to HDFS. One with ten records and was used for quick testing, and two others were used for the actual benchmarking of 50 million records and one billion records. The measurements were automated as much as possible to make easy for the eventual benchmarking of 10 billion records. Figure 6 shows the fluxes of the IE for all 60 iterations to three billion. We also computed the ingestion efficiency (IE) of one billion compared to 50 million records using the formula in equation (1):
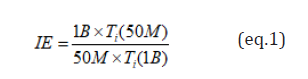
where Ti (N) is the time it takes to ingest N records to either HDFS or HBase.
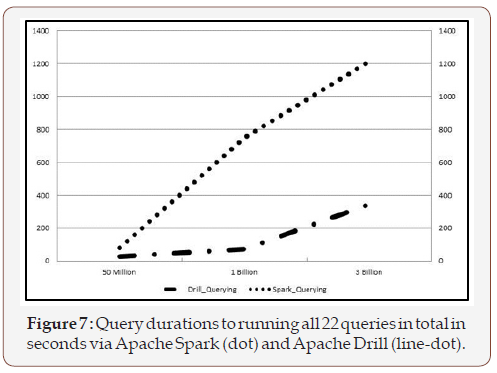
A SQL script containing all the queries was written and ran using Phoenix sqlline.py. The output of sqlline.py was redirected to a file for further validation. In addition to the query results, sqlline. py reported the time it took for each query (similar to the usual SQL query clients). The total number of queries that were used was 22: two simple queries with wildcard column selection; ten simple queries that did not involve more than three columns in the primary keys (family qualifiers); and, ten complex queries that had >3 columns selected. These queries were chosen based on surveys and interviews with VIHA experts on current clinical reporting. Furthermore, the separation between simple and complex queries was also derived to compare to performance of Apache Spark and Apache Drill on the exact same data (Figure 7).
Discussion
Essentially this study is proposing a row-column key-value (KV) model to hold the Hadoop/MapReduce to the data distributed over a customized BDA platform for healthcare application. Wang, Goh, Wong, and Montana [54] support this study’s claim in their statement that non-relational data models, such as the KV model implemented in NoSQL databases exhibit accurate multivariate analysis of Big Data in Hadoop forest using data from bioinformatics. Wang et al. [55] further stated that NoSQL provided high performance solutions for healthcare, being better suited for high-dimensional data storage and querying, optimized for database scalability and performance. A KV pair data model can support faster queries of large-scale microarray data and can be implemented using HBase, i.e., an implementation of Google’s Bigtable storage system. This new KV data model implemented on HBase exhibited an average 5.24-fold increase in highdimensional biological data query performance compared to the relational model implemented on MySQL Cluster and an average 6.47-fold increase on query performance on MongoDB [40]. The performance evaluation found that the new KV data model, in particular its implementation in HBase, outperforms the relational model currently implemented, and, therefore, supports this study’s NoSQL technology for large-scale data operational BDA platform of hospital systems.
With HBase, the table schema is dynamic because it can be modified at any time and incorporates other forms of data for different schemas [56]. However, in order for the platform to function, HBase’s schema must be predefined (via SQL-like Phoenix in this study), and the column families specified before it is operational. However, the schema is very flexible, in that new columns can be added to families or groups at any time. It is therefore able to adapt to changing application requirements, as has been noted by many researchers [57, 58]. As Sun [59] states, “Just as HDFS has a Name Node and slave nodes, and MapReduce has Job Tracker and Task Tracker slaves, in HBase a master node (HMaster) manages the cluster and Region Servers store portions of the tables and perform the work on the data.” HMaster is the implementation of the Master Server, which is responsible for monitoring all Region Server instances in the cluster. In this study’s distributed cluster, the Master started on the Name Node, while HRegion Server started the Region Server(s). In a distributed cluster, a Region Server runs on a Data Node [59], as was the case in our implementation. In HBase, through the Zookeeper, other machines can be selected within the cluster as HMaster (unlike the HDFS architecture, in which Name Node has a single-point-of-availability problem) [59]. HBase clusters can also be expanded by adding Region Servers hosted on commodity class servers. For example, when a cluster expands from 10 to 20 Region Servers, it doubles both in terms of storage and processing capacity. Therefore, more storage space will be required when the number of nodes increases. The present study did confirm that HBase and HDFS worked efficiently; HBase supported parallelized processing via MapReduce; and the platform provided a user-friendly Java API for programmatic enduser access.
It is possible to mimic a relational database with an SQLlike structure using NoSQL [52]. The primary key strength of the patient encounter was created using “family” or groups of columns combined. And this combination proved necessary to show accurate query results on real patient data when utilized. Without the groupings to form the SQL-like primary key, the results from the query did not show the correct number of fields found. In addition, the key values must be unique; therefore, when replicating the ingestion, queries had to be modified; otherwise no result could be obtained. Chawla and Davis [60] emphasize the importance of the patient-centered framework for the Big Data platform. Since many Big Data platforms are linked to HDFS with non-relational databases, assuring the accuracy of patient data is of the utmost importance. This was achieved in simulating queries over NoSQL database but not fully validated over with non-replicated data of more detailed patient-centric encounters.
Even though it is very difficult to move the patient data from the hospital system that maintains its clinical model stored in real-time, analyzing patient-level databases can yield populationlevel inferences or “results” (such as the strength of association between medical treatments and outcomes), often with thousands of outcomes. Other studies indicate that although data from such sources as hospital EHR systems are generally of much lower quality than data carefully collected by researchers investigating specific questions, the sheer volume of data can compensate for its qualitative deficiencies, provided that a significant pattern can be found amid the noise [3,61]. In addition, there is a trend toward higher quality Big Data collections (such as the data produced in genomic analysis and structured data generated from standardcompliant EHR systems) [62,63]. On the other hand, Mayer- Schonberger and Cukie [64] showed that as the percentage of the population being sampled approaches 100%, messy data can have greater predictive power than highly cleaned and carefully collected data that might represent only a small sample of the target population. The present study did not analyze the structure of the metadata and ultimately it was designed to replicate massive volumes of patient data.
It was more complicated to validate the simulated data in Spark and Drill to validate a proof-of-concept use of these tools with real data. Scott [55] indicated that the strategies for the best big data software solutions is between Spark and Drill and that Drill can emulate complex data much more efficiently than Spark because Spark requires elaborate Java, Python and Scala coding. Nonetheless, both Spark and Drill were significantly faster than HBase in ingesting files directly into Hadoop. In fact, the ingestion and queries for both Spark and Drill could be run in sequence instead of having to run compaction as well. However, it is difficult to compare the data emulation of HBase to that of Spark and Drill. Neither Spark nor Drill indexed the files, nor did they fully require the Reducer from MapReduce to complete its task before queries could be performed. Absence of indexing increases the risk of inaccuracies (even though the framework was more faulttolerant when running Spark and Drill). Therefore, the big data tools and inherent technologies therein highly influence the health informatics of the data established and resulting queries.
The most impactful technology in this study was MapReduce (and its Java code). MapReduce methodology is inherently complex as it has separate Map and Reduce task and steps in its default programming framework as this study discovered. This study’s platform was highly dependent on the efficiency of MapReduce in ingesting files over the six nodes, using this workflow: Input Map → Copy/Sort → Reduce → Output similar to a study by Chen, Alspaugh and Katz [65]. Once configurations in Yarn, ZooKeeper, and others the Reducer were optimized with iterations of 50 million rows with data, integrity of the desired clinical event model was established via SQL-like in Apache Phoenix. According to blogs with technical resolutions, enabling or disabling services or xml settings over the platform is expected to be carried out because the system relies heavily on InfiniBand (IB) bandwidth and not all default settings can be applied automatically when initializing. Furthermore, there other known issues with using MapReduce over HBase with slow performance after additional indexing of data and its store [38,40,66-68].
The data used in this study consisted of diagnosis codes, personal health numbers, medical record numbers, dates of birth, and location mnemonics (to mention only a few of the 90 columns), as these codes are standardized for hospital systems and, compared to genetic data, easier to replicate metadata in large volumes. The use of groups of events allowed the definition of a phenotype to go beyond diagnosis as coded using the International Classification of Disease, version 9, codes (ICD-9) and potentially allows assessment of the accuracy of assigned codes [62,69]. In healthcare, the complexity of Big Data storage and querying increase with unstructured sets of data and/or images. The growth in the volume of medical images in modern hospitals has forced a move away from traditional medical image analysis and indexing approaches towards scalable solutions [70]. In fact, MapReduce has been used to speed up and make possible three large-scale medical image processing use-cases: (1) parameter optimization for lung texture classification using support vector machines (SVM), (2) contentbased medical image indexing/retrieval, and (3) dimensional directional wavelet analysis for solid texture classification [71]. In their study, a default cluster of heterogeneous computing nodes was set up using the Hadoop platform, allowing for a maximum of 42 concurrent Map tasks. However, this study did not test the amount and efficiency of concurrent Map tasks of MapReduce to process the data to HBase ingestions.
Greeshma & Pradeepini [66] implemented a distributed file system that stored Big Data files, and a MapReduce algorithm supported the performance of the analytics on a set of clusters. The platform queries did not rely on MapReduce to perform and only controlled the actual ingestion of files to form the database. Spark and Drill can use some components of MapReduce (or limited usage) at the initialization of the queries, but this was minimal and non-reliable. The Hadoop approach used in this study’s platform did not consume data on a large scale as a whole but broke the computational data into smaller pieces over the collection of servers and iterations. This placed the bulkloading on the AdminNode (deployment) that was deployed to NameNode and then distributed to DataNode. Our findings support Greeshma and Pradeepini’s [66] findings that NameNode and AdminNode usually manage everything with respect to storing, managing, and accessing Big Data files.
This study did not benchmark the actual performance of the components of the platform to the existing system that uses analytical tools across EHR. However, to replicate/transfer data from production to domains in the hospital system via secure FTP takes six or more months at 10-15 TB. Thus, for our dataset volumes 30-40 TB over three months could be in the order of ten-fold magnitude faster than existing technical platform. Moreover, Yu et al. [68] showed that Mahout’s MapReduce architecture was eight to five times slower than other modified MapReduce configurations. More specific to our proposed framework, Greeshma & Pradeepini [66] showed that adding Job Tracker to the MapReduce framework is useful, since Hadoop does need to be running while doing the queries; it solves the limitations of the relational data warehouse by continuously running with low resources over multiple nodes. Hence, Hadoop is a single point of failure since it cannot stop running to perform any tasks or functionalities, which is supported findings of broke Hadoop clusters by Rabkin & Katz [72].
A large body of research has studied ways of improving storage performance [73] or using compression techniques to reduce the volume of intermediate data [74,75]. The intermediate data movement time during the shuffle phase has been addressed [76,77]. However, none of these studies addressed the issue of memory limitation in the Reduce phase, which can lead to abortion of Reduce tasks, as reported by Yan et al. [78], who indicated the importance of memory resources (which is exactly what occurred with ingestions in this study, although the platform did not fail any time queries were run). Greeshma & Pradeepini [66] did propose a memory cached mechanism to utilize with MapReduce to improve the Reducer, but this was not tested on other software technologies like HBase. Additionally, while saturation of storage IO operations or network bandwidth can lead to performance degradation in the Reduce phase (a problem encountered on Regional Servers), the MapReduce application cannot be killed by the Hadoop framework in such cases [79]. However, in the case of an out-of-memory error, the job is usually killed, since the Reduce phase needs to bring large portions of intermediate data into memory for processing [80].
The present study showed that performing maintenance activities over the platform were essential for ensuring reliability. Some studies have shown that Hadoop can detect task failure and restart programs on healthy nodes, but if the RegionServers for HBase fail, this process had to be started manually [67,81,82]. Our study showed that compaction improved the number of successful runs of ingestion; however, it did not prevent failure of ingesting the files, a finding that is also supported by other studies [36,38,80,83].
Conclusion
Few studies have produced a healthcare BDA platform and this study adds to Big Data technologies, data simulation and healthcare applications over large volumes. This study proposes a framework that not only replicates large amounts of data over platform but allows for combining data from different databases at each generation of the replication. The results showed that to replicate data from source to HBase to form three billion patient encounters required a month’s timeframe; therefore, the data needs to be stored before running the queries. Further, Moselle [84] states that if data is stored with some permanency over a certain time period longer than a few hours without removal, public disclosure is required (in accordance with privacy/security laws related to personal sensitive data). Furthermore, Dufresne, Jeram, and Pelletier [85] point out that Canadians have a nationalistic view of their healthcare and having public data with publicly provided healthcare makes sense to many citizens, including providers and health professionals. Thus, if the BDA platform can produce accurate query results within seconds while the data is either still housed in the hospital or encrypted and highly secure then either public disclosure can be deemed not necessary or change the policy.
Useful knowledge gained from this study included the following challenges:
• data aggregation - actual storage doubled compared to what was expected due to HBase key store qualifiers, and big datasets of clinical, biomedical, and biometric data have been processed successfully with Hadoop/MapReduce framework;
• data maintenance – ingestions to the database required continual monitoring and updating versions of Hadoop (HDFS), MapReduce, and HBase with limitations persisting for MapReduce;
• data integration – combination of ADT and DAD is possible and followed current clinical reporting but requires a more complex clinical event model of the row keys and column families to be further tested;
• data analysis – high performance of 3.5 seconds for three billion rows and 90 columns (30TB) achieved with increasing complexity of queries; and
• pattern interpretation of application – health trends cannot be found via the application and further investigation required using Hadoop’s Machine Learning Libraries (MLLib).
The design of the implemented BDA platform (utilizing WestGrid’s supercomputing clusters) is available to researchers and sponsored members. The next step of the testing of the BDA platform will be to distribute and index the data to ten billion patient data rows across the database nodes, and then test the performance using the established methodology. Afterwards, the study plan is to distribute the data across 100 nodes; Hadoop’s HDFS and HBase are theoretically supposed to scale and function better with more nodes [14,15].
References
- M Chen, S Mao, Y Liu (2014) Big Data: A Survey. Mobile Network Application 19(2): 171-209.
- M Viceconti, P Hunter, R Hose (2015) Big data, big knowledge: big data for personalized healthcare. IEEE Journal of Biomedical and Health Informatics 19(4): 1209-1215.
- MM Hansen, T Miron Shatz, AYS Lau, C Paton (2014) Big Data in Science and Healthcare: A Review of Recent Literature and Perspectives, Contribution of the IMIA Social Media Working Group. Yearbook of Medical Informatics 9(1): 21-26.
- J Manyika, M Chui, J Bughin, B Brown, R Dobbs, C Roxburgh, B Hung (2011) Big data: The next frontier for innovation, competition, and productivity. p. 1-137.
- Canada Health Infoway, Big Data Analytics in Health - White Paper, 2013.
- W Raghupathi, V Raghupathi (2014) Big data analytics in healthcare: promise and potential. Health Information Science and Systems 2(3): 1-10.
- R. Foster, Health Care Big Data is a big opportunity to address data overload.
- NH Shah, Tenenbaum JD (2012) The coming age of data-driven medicine: translational bioinformatics’ next frontier. Journal of the American Medical Informatics Association 19: e2-e4.
- LP Jr Garrison (2013) Universal Health Coverage-Big Thinking versus Big Data. Value Health 16(1): S1-S3.
- E Baro, S Degoul, R Beuscart, E Chazard (2015) Toward a literature-drive definition of big data in healthcare. BioMed Research Inter-national p. 9.
- B Wang, R Li, W Perrizo (2014) Big Data Analytics in Bioinformatics and Healthcare. Medical Information Science Reference p. 459.
- MH Kuo, T Sahama, AW Kushniruk, EM Borycki, D Grunwell (2014) Health Big Data Analytics: Current Perspectives, Challenges and Potential Solutions. International Journal of Big Data Intelligence 1(4): 114-126.
- J Seidman, G Shapira, T Malaska, M Grover (2015) Hadoop: Application Architectures-Designing Real-World Big Data Applications. O’Reilly Media, USA, pp. 1-364.
- T White (2015) Hadoop: The Definitive Guide: Storage and analysis at internet scale (4th edn). O’Reilly Publishing, USA, pp. 1-235.
- WK Lai, YC Chen, Tin Yu Wu, MS Obaidat (2014) Towards a framework for large-scale multimedia data storage and processing on Hadoop platform. J Supercomp 68(1): 488-507.
- L Dai, X Gao, Y Guo, J Xiao, Z Zhang (2012) Bioinformatics clouds for big data manipulation. Biology Direct 7(43): 1-7.
- A Kumar, S Bawa (2012) Distributed and big data storage management in grid computing. International Journal of Grid Computing and Applications 3(2): 19-28.
- I Merelli, H Perez Sanchez, S Gesing, DD Agostino (2014) Managing, Analysing, and Integrating Big Data in Medical Bioinformatics: Open Problems and Future Perspectives. BioMed Research International pp. 1-14.
- ABM Moniruzzaman, SA Hossain (2013) NoSQL Database: New Era of Databases for Big Data Analytics - Classification, Characteristics and Comparison. International Journal of Database Theory and Application 6(4): 1-14.
- A Lith, J Mattson (2010) Investigating storage solutions for large data-A Comparison of well performing and scalable data storage solutions for real time extraction and batch insertion of data. Chalmers University of Technology, Sweden.
- E Pattuk, M Kantarcioglu, V Khadilkar, H Ulusoy, S Mehrotra, (2013) Big Secret: A Secure Data Management Framework for Key-Value Stores. IEEE Sixth International Conference on Cloud Computing pp. 147-154.
- P Derbeko, S Dolev, E Gudes, S Sharma (2016) Security and Privacy Aspects in MapReduce on Clouds: A Survey. Computer Science Review 20: 1-28.
- F Martin Sanchez, K Verspoor (2014) Big Data in Medicine Is Driving Big Changes. Yearbook of Medical Informatics 9(1): 14-20.
- M Kwakye (2011) A Practical Approach to Merging Multidimensional Data Models. Master thesis, School of Electrical Engineering and Computer Science, University of Ottawa, Canada.
- J Chen, Y Chen, X Du, C Li, J Lu, S Zhao et al. (2013) Big data challenge: a data management perspective. Frontiers of Computer Science 7(2): 157- 164.
- NJ Leeper, A Bauer Mehren, SV Iyer, P Lependu, C Olson et al. (2013) Practice-based evidence: profiling the safety of cilostazol by text-mining of clinical notes. Plos One 8(5): 1-8.
- P Lependu, SV Iyer, C Fairon, NH Shah (2012) Annotation analysis for testing drug safety signals using unstructured clinical notes. Journal of Biomedical Semantics 3(1): S1-S5.
- W Raghupathi, V Raghupathi (2014) Big data analytics in healthcare: promise and potential. Health Information Science and Systems 2(3): 1-10.
- EE Schadt, MD Linderman, J Sorenson, L Lee, GP Nolan (2010) Computational solutions to large-scale data management and analysis. Nature Reviews 11: 647-657.
- K Deepthi, K Anuradha (2016) Big data Mining Using Very-Large-Scale Data Processing Platforms. International journal of engineering research and applications 6(2): 39-45.
- A Hashmi, T Ahmad (2016) Big Data Mining: Tools & Algorithms. International Journal of Recent Contributions from Engineering, Science & IT 4(1): 21.
- YP Zhang, Shimin Chen, Qiang Wang, Ge Yu (2015) i2 MapReduce: Incremental MapReduce for Mining Evolving Big Data. IEEE Transactions on Knowledge and Data Engineering 27(7): 1906-1919.
- R Narawade, V Jadhav (2015) Data Mining of Big Data: The Survey and Review. International Journal of Innovations in Engineering and technology 6(2): 248-252.
- DP Vaidya, SP Deshpande (2015) Parallel Data Mining Architecture for Big Data. International Journal of Electronics, Communication and Soft Computing Science and Engineering, pp. 208-213.
- X Wu, X Zhu, GQ Wu, W Ding (2014) Data Mining with Big Data. IEEE Transactions on Knowledge and Data Engineering 26(1): 97-107.
- EA Mohammed, BH Far, C Naugler (2014) Applications of the MapReduce programming framework to clinical big data analysis: current landscape and future trends. BioData Mining 7(22): 1-23.
- F Marozzo, D Talia, P Trunfio (2012) P2P-MapReduce: Parallel data processing in dynamic cloud environments. Journal of Computer and System Sciences 78(5): 1382-1402.
- RC Taylo (2010) An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC Bioinformatics 11(12): 1-6.
- P Langkafel (2016) Big Data in Medical Science and Healthcare Management: Diagnosis, Therapy, side Effects. De Gruyter, ISBN: 3110445751.
- S Sakr, Amal Elgammal (2016) Towards a comprehensive data analytics framework for smart healthcare services. Big Data Research 4: 44-58.
- RS Chang, CS Liao, KZ Fan, CM Wu (2014) Dynamic Deduplication Decision in a Hadoop Distributed File System. International Journal of Distributed Sensor Networks, pp. 1-14.
- LB Madsen (2014) Data-Driven healthcare: how analytics and BI are transforming the industry. Jon Wiley and Sons ISBN: 9781118772218.
- W Raghupathi, V Raghupathi (2014) Big data analytics in healthcare: promise and potential. Health Information Science and Systems 2(3): 1-10.
- ZooKeeper (2016) ZooKeeper - Apache Software Foundation
- K Sitto, M Presser (2015) Field Guide to Hadoop - An Introduction to Hadoop, Its Ecosystem, and Aligned Technologies. O Reilly Media, San Francisco, CA, USA.
- T Dunning, E Friedman, T Shiran, J Nadeau (2006) Apache Drill. O Reilly Media, San Francisco, CA, USA.
- R Jurney (2013) Agile Data Science: Building Data Analytics Applications with Hadoop. O Reilly Media, San Francisco, CA.
- WestGrid (2016).
- J Benaloh, M Chase, E Horvitz, K Lauter (2009) Patient Controlled Encryption: Ensuring Privacy of Electronic Medical Records. Proc. ACM Workshop on Cloud Computing Security, USA, pp. 103-114.
- D Chrimes, MH Kuo, B Moa, W Hu (2016) Towards a Real-time Big Data Analytics Platform for Health Applications. Int. J. Big Da-ta Intel 4(2).
- D Chrimes, B Moa, H Zamani, MH Kuo, (2016) Interactive Healthcare Big Data Analytics Platform under Simulated Performance. IEEE 14th Int. Conf. Dependable, Autonomic and Secure Computing, Pervasive Intelligence and Computing, 2nd Intl. Conf. on Big Data Intelligence and Computing and Cyber Science and Technology Congress, pp. 811-818.
- J Xu, M Shi, C Chen, Z Zhang, J Fu, et al. (2016) ZQL: A unified middleware bridging both relational and NoSQL databases. IEEE 14th Int. Conf. Dependable, Autonomic and Secure Computing, Pervasive Intelligence and Computing, 2nd Intl. Conf. on Big Data Intelligence and Computing and Cyber Science and Technology Congress, pp. 730-737.
- CT Yang, JC Liu, WH Hsu, WCC Chu (2013) Implementation of data transform method into NoSQL database for healthcare data. International Conference on Parallel and Distributed Computing, Applications and Technologies.
- Y Wang, W Goh, L Wong, G Montana (2013) Random forests on Hadoop for genome- wide association studies of multivariate neuroimaging phenotypes. BMC Bioinformatics 14(16): 1-15.
- S Wang, I Pandis, C Wu, S He, D Johnson et al. (2014) High dimensional biological data retrieval optimization with NoSQL technology. BMC Genomics 15(8): S3.
- J Scott (2015) Apache Spark vs. Apache Drill. Converge Blog, Powered by MapR.
- S Nishimura, S Das, D Agrawal, AE Abbadi (2012) MD-HBase: design and implementation of an elastic data infrastructure for cloud-scale location services, Springer Science+Business Media, LLC.
- AV Nguyen, R Wynden, Y Sun (2011) HBase, MapReduce, and Integrated Data Visualization for Processing Clinical Signal Data. AAAI Spring Symposium: Computational Physiology, pp. 40-44.
- J Sun (2013) Scalable RDF store based on HBase and MapReduce. Advanced Computer Theory and Engineering (ICACTE), 3rd Int. Conf., Hangzhou, China.
- NV Chawla, DA Davis (2013) Bringing Big Data to Personalized Healthcare: A Patient- Centered Framework. J Gen Intern Med 28 (3): S660-S665.
- PO Sullivan, G Thompson, A Clifford (2014) Applying data models to big data architectures. IBM J Res & Dev 58(5): 1-12.
- SM Freire, D Teodoro, F Wei Kleiner, E Sundsvall, D Karlsson et al. (2016) Comparing the Performance of NoSQL Approaches for Managing Archetype-Based Electronic Health Record Data. PLoS One 11(3).
- LJ Frey, L Lenert, G Lopez Campos (2014) EHR Big Data Deep Phenotyping Contribution of the IMIA Genomic Medicine Working Group. Year Med Inform 9: 206-211.
- V Mayer Schonberger, K Cukie (2013) Big data: A revolution that will transform how we live, work, and think, Houghton Mifflin Harcourt. Chapter 2.
- Y Chen, S Alspaugh, R Katz (2012) Interactive Analytical Processing in Big Data Systems: A Cross-Industry Study of MapReduce Workloads. Proceedings of the VLDB Endowment 5(12): 1802-1813.
- AL Greeshma, G Pradeepini (2016) Input split frequent pattern tree using MapReduce paradigm in Hadoop. J Theo App Inform Tech 84(2): 260-271.
- M Maier (2013) Towards a Big Data Reference Architecture. MSc Thesis. Eindhoven University of Technology, Netherlands, pp. 1-144.
- SC Yu, QL Kao, CR Lee (2016) Performance Optimization of the SSVD Collaborative Filtering Algorithm on MapReduce Architectures. IEEE 14th Int. Conf. Dependable, Autonomic and Secure Computing, Pervasive Intelligence and Computing, 2nd Intl Conf on Big Data Intelligence and Computing and Cyber Science and Technology Congress, pp. 612-619.
- Hripcsak, G & Albers, DJ (2013) Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 20(1): 117-21.
- F Wang, R Lee, Q Liu, Ablimit Aji, X Zhang, J Saltz (2011) Hadoop-GIS: A high performance query system for analytical medical imaging with MapReduce. Emory University, pp. 1-13.
- D Markonis, R Schaer, I Eggel, H Muller, A Depeursinge (2012) Using MapReduce for large-scale medical image analysis. Healthcare Informatics, Imaging and Systems Biology (HISB), IEEE Second International Conf. La Jolla, CA, USA, September 27-28, pp. 1-10.
- A Rabkin, RH Katz (2013) How Hadoop Clusters Break. IEEE Software 30(4): 88-95.
- D Moise, TTL Trieu, L Bouge, G Antoniu (2011) Optimizing intermediate data management in MapReduce computations. Pro-ceedings of the first international workshop on cloud computing platforms, ACM, pp. 1-7.
- G Ruan, H Zhang, B Plale (2013) Exploiting MapReduce and data compression for data-intensive applications. Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery, ACM, pp. 1-8.
- Z Xue, G Shen, J Li, Q Xu, Y Zhang, J Shao (2012) Compressionaware i/o performance analysis for big data clustering. Proceedings of the 1st International Workshop on Big Data, Streams and Heterogeneous Source Mining: Algorithms, Systems, Programming Models and Applications, ACM, pp. 45-52.
- Y Wang, C Xu, X Li, W Yu (2013) JVM-bypass for efficient Hadoop shuffling. In: IEEE 27th International Symposium on Parallel & Distributed Processing (IPDPS), pp. 569-578.
- W Yu, Y Wang, X Que, C Xu (2015) Virtual shuffling for efficient data movement in MapReduce. IEEE Transactions on Computers 64(2): 556- 568.
- D Yan, XS Yin, C Lian, Xiang Zhong, Xin Zhou et al. (2015) Using memory in the right way to accelerate big data processing. Journal of Computer Science and Technology 30(1): 30-41.
- F Ahmad, ST Chakradhar, A Raghunathan, T Vijaykumar (2014) Shufflewatcher: Shuffle-aware scheduling in multi-tenant MapReduce clusters. Proceedings of the USENIX conference on USENIX Annual Technical Conference. USENIX Association, pp. 1-12.
- SM Nabavinejad, M Goudarzi, S Mozaffari (2016) The Memory Challenge in Reduce Phase of MapReduce Applications. 14(8): 380-386.
- WC Chung, HP Lin, SC Chen, MF Jiang, YC Chung (2014) JackHare: a framework for SQL to NoSQL translation using MapReduce. Autom Softw Eng 21(4): 489-508.
- H Dutta, Alex Kamil, Manoj Pooleery, Simha Sethumadhavan, J Demme (2011) Distributed Storage of Large Scale Multidimensional EEG Data using Hadoop/HBase. Grid and Cloud Database Management, New York City, Springer, pp. 331-347.
- J Dean, S Ghemawat (2010) MapReduce: A Flexible Data Processing Tool. Comm ACM 53(1): 72-77.
- K Moselle (2015) Data Management in the Island Health Secure Research Environment. Enterprise Architecture at Vancouver Island Health Authority, Working Draft 5, Victoria, BC.
- Y Dufresne, S Jeram, A Pelletier (2014) The True North Strong and Free Healthcare? Nationalism and Attitudes Towards Private Healthcare Options in Canada. Canadian Journal of Political Science 47(3): 569-595.
-
D Chrimes, MH Kuo, AW Kushniruk, B Moa. Interactive Big Data Analytics Platform for Healthcare and Clinical Services. Glob J Eng Sci. 1(1): 2018. GJES.MS.ID.000502.
-
Analytics platform, Healthcare and clinical services, Adaptable architectures, Big data, Data mining, Distributed filing system, Distributed data structures, Healthcare informatics, Hospital systems, Metadata, Relational database, Data maintenance, Data aggregation, Data integration, Data analysis, Data maintenance, Pattern interpretation, Data emulation and modeling, Computing platform and configurations, The data process pipeline
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






