 Short communication
Short communication
Pharmacogenetic Clinical Decision Support in Patients with Multi-Drug Regimens
Jorge Duconge1 and Gualberto Ruaño2*
1School of Pharmacy, Medical Sciences Campus, University of Puerto Rico, San Juan, PR USA
2Institute of Living at Hartford Hospital, Hartford and Department of Psychiatry, Univ. of Connecticut School of Medicine, Farmington, CT USA
Gualberto Ruaño, Institute of Living at Hartford Hospital, Hartford, CT, USA.
Received Date: September 16, 2022; Published Date: November 17, 2022
Introduction
We had reported our experience in a comprehensive medication management service for mental health with an algorithmic and heuristic Clinical Decision Support system (MEDtuning, Genomas Inc.) [1] based on the combinatorial genetic profile of CYP2D6, CYP2C19 and CYP2C9 [2]. This system provides a drug proscriptive warning if a patient’s CYP450 phenotype (derived from the genotypes) is functionally abnormal for an isoenzyme constituting the primary or sole metabolic pathway for a given medication. The CDS tool has previously proven very useful for diagnosing pharmacogenetic vulnerabilities of psychiatric patients and supporting drug selections to minimize risk [3-5]. Multidrug regimens (i.e., polypharmacy) have the greatest potential for bringing drug-related adverse events and harmful drug interactions in patients with cardiometabolic and neuropsychiatric co-morbidities.
Table 1:CDS results from 2,470 US patients of different ethno-geographic regions. The first 2 columns classify the cohort by number of different drugs prescribed and the number of patients receiving each amount. The italicized columns indicate the prescription of high-risk drugs, and the number of patients receiving these for each of the drug regimens. The last column provides the percentage of patients in each of the drug regimens receiving at least 1 high risk drug.
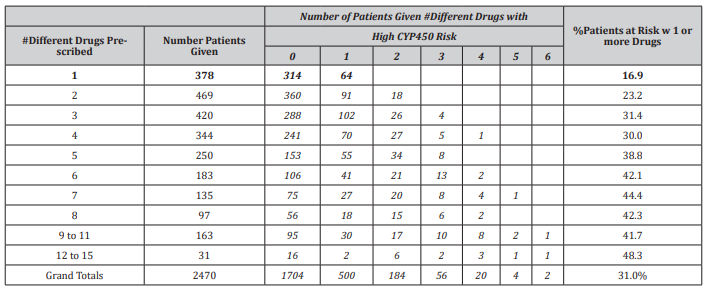
[Table 1] Illustrates the application of the pharmacogenetic CDS to 2,470 patients of diverse ethno-geographic backgrounds (including Hispanics) referred to the clinical laboratory. Nearly half of the patients (49%) was prescribed 4 or more drugs. The number of patients given high-risk drugs identified by the CDS system steadily increased as the number of prescribed medications rose. The percentage ranged from 17% of patients being given a highrisk drug when only one medication was prescribed up to 48.3% of patients being given high risk drug(s) when given 12-15 medications. Altogether, 31% of patients in the cohort had been prescribed at least 1 high risk medication.
In the future, it should be possible for the pharmacist to apply the genotype data to selected CDS models from various sources and achieve a consensus based on clinical judgment. This proscriptive global model could complement specific clinical guidance’s available for selected drug-gene pairs from learned bodies [6-9]. These models can enable comprehensive medication management, as no single pharmacogenetic guidance model is likely to encompass the multiple drug regimens overseen by the pharmacy profession.
Acknowledgement
Patients signed an informed consent agreeing to DNA testing and use of deidentified information for validation, research, and accreditation purposes. This project was supported by Genomas internal research funds.
Conflict of interest
GR was Medical Director of Genomas when patients were referred.
References
- Rodríguez Escudero I, Cedeño JA, Rodríguez Nazario, Reynaldo Fernandez G, Rodríguez Vera L, et al. (2020) Assessment of the clinical utility of pharmacogenetic guidance in a comprehensive medication management service. J Am Coll Clin Pharm 3(6): 1028-1037.
- Villagra D, Goethe J, Schwartz HI, Szarek B, Kocherla M, et al. (2011) Novel drug metabolism indices for pharmacogenetic functional status based on combinatory genotyping of CYP2C9, CYP2C19 and CYP2D6 genes. Biomark Med 5(4): 427-38.
- Ruaño G, Blair CL, Bower B, Windemuth A, Kocherla M, et al. (2007) Somatic complications of psychotropic medications in a patient with multiple CYP2 drug metabolism deficiencies. Connecticut medicine71(4): 197-200.
- Landino J, Buckley J, Roy JM, Villagra D, Gorowski K, et al. (2011) Guidance of pharmacotherapy in a complex psychiatric case by CYP450 DNA typing. Journal of the American Academy of Nurse Practitioners23(9): 459-463.
- Ruaño G, Larsen K, Kocherla M, Graydon JS, Kost J (2017) Complications of psychotropic and pain medications in an ultrarapid metabolizer patient at the upper 1% of cytochrome P450 (CYP450) function quantified by combinatorial CYP450 genotyping. Journal of pain and palliative care pharmacotherapy 31(2): 126-38.
- Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, et al. (2013) Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clinical Pharmacology and Therapeutics 93(5): 402-408.
- Crews K R, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, et al. (2014) Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy 2014 update. Clinical Pharmacology and Therapeutics 95(4): 376-382.
- Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, et al. (2015) Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clinical Pharmacology and Therapeutics 98(2): 127-134.
- Jessel CD, Mostafa S, Potiriadis M, Everall IP, Gunn JM, et al. (2020) Use of antidepressants with pharmacogenetic prescribing guidelines in a 10-year depression cohort of adult primary care patients. Pharmacogenetics and Genomics 30(7): 145-52.
-
Jorge Duconge and Gualberto Ruaño*. Pharmacogenetic Clinical Decision Support in Patients with Multi-Drug Regimens. Curr Tr Clin & Med Sci. 3(2): 2022. CTCMS.MS.ID.000560.
-
Psychiatric patients, Pharmacogenetic, CDS models, clinical laboratory, patients
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






