 Research Article
Research Article
Investigating the Role of Cannabinoids from Cannabis sativa in Pancreatic Beta Cell Differentiation via WNT5A/JNK and BMP Pathways: An In-Silico Approach
Guy Roussel Takuissu1,2*, Shabnor Iqbal1, Cedric Fossi Tchinda1,2, Fils Armand Ella1,3, Corinne Raissa Ngnameko1,2, Matsabisa Gilbert Motlalepula1*
1Department of Pharmacology, University of the Free State, Bloemfontein, South Africa
2Institute for the Medical Research and Medicinal Plant Studies (IMPM), Ministry of Scientific Research and Innovation, Yaoundé, Cameroon
3Department of Biochemistry, University of Yaounde, Yaounde, Cameroon
Matsabisa Gabriel Motlalepula, Department of Pharmacology, University of the Free State, Bloemfontein, South Africa Guy Roussel Takuissu, Department of Pharmacology, University of the Free State, Bloemfontein, South Africa
Received Date:January 16, 2025; Published Date:February 18, 2025
Abstract
Pancreatic beta cell dysfunction is a hallmark of diabetes, and targeting pathways regulating their differentiation holds promise for novel therapies. This study aims to investigate the role of cannabinoids from Cannabis sativa in pancreatic beta cell differentiation via the WNT5A/ JNK and BMP pathways using an in-silico approach. The study employed molecular docking simulations to predict the study employed molecular docking simulations to predict the binding affinity and molecular interactions of six cannabinoids with key proteins involved in the WNT5A/JNK (WNT5A, JNK, c-JUN, and FZD3) and BMP (BMP6, SMAD 1, 4 and 5) pathways. The stability and accuracy of the protein-ligand complexes obtained from molecular docking simulations were validated using RMSD values, and normal mode analysis was conducted to gain insights into molecular flexibility and deformation. RMSD analysis validated the stability of predicted binding modes, and normal mode analysis was conducted to gain insights into molecular flexibility and deformation.
The molecular docking results showed that all six cannabinoids had binding affinities with proteins involved in the WNT5A/JNK and BMP pathways, with the highest affinity observed for the JNK-Cannabinol and SMAD4- tetrahydrocannabivarin complex. The RMSD values validated the accuracy and stability of the protein-ligand complexes. Specific interactions, such as hydrophobic contacts and hydrogen bonds, were identified between JNK-Cannabinol and SMAD4- tetrahydrocannabivarin. Normal mode analysis revealed flexible and rigid regions within the complexes that could impact interaction surfaces. The findings of this study suggest that cannabinoids may directly impact beta cell differentiation signaling cascades and warrant further investigation into their potential as anti-diabetic therapies.
Keywords:Cannabinoids; pancreatic beta cell differentiation; WNT5A/JNK pathway; BMP pathway; in silico approach
Introduction
The therapeutic potential of Cannabis sativa has been a subject of scientific intrigue for decades, with growing evidence suggesting that its cannabinoids have the ability to influence cellular pathways and modulate various physiological processes [1]. Among the many therapeutic applications of cannabinoids, their potential role in pancreatic beta cell differentiation has gained considerable attention in recent years. Pancreatic beta cells are responsible for the production and secretion of insulin, and their dysfunction or loss is a hallmark of diabetes [2]. Therefore, understanding the molecular mechanisms underlying beta cell differentiation and identifying novel therapeutic targets for diabetes treatment is of utmost importance. The WNT5A/JNK and BMP pathways are known to play critical roles in pancreatic beta cell differentiation and function [3,4].
In particular, the BMP pathway has been shown to promote beta cell proliferation and differentiation, while the WNT5A/JNK pathway has been implicated in regulating beta cell survival and function [5,6]. In the BMP pathway, BMP4/6 bind to their receptors and activate SMAD 1/4/5, which then translocate to the nucleus and regulate gene expression [7]. In the WNT5A/JNK pathway, WNT5A binds to its receptor FZD3, leading to the activation of JNK and c-JUN, which regulate gene expression and cellular processes [8]. Dysregulation of these pathways has been linked to the development of diabetes, highlighting the need to understand the molecular mechanisms underlying their regulation and to identify novel therapeutic targets for diabetes treatment.
Emerging evidence suggests that cannabinoids can modulate these pathways, potentially influencing beta cell differentiation and survival [9,10]. However, the molecular mechanisms underlying the effects of cannabinoids on these pathways remain largely unknown. Previous studies have focused on the antidiabetic activities of cannabinoids, primarily through their effects on insulin sensitivity and inflammation [11,12]. While these studies have provided valuable insights into the potential therapeutic benefits of cannabinoids in diabetes, the direct effects of cannabinoids on beta cell differentiation and function have not been fully explored. This study aims to address this gap in knowledge by investigating the role of cannabinoids from Cannabis sativa (cannabidiol (CBD), tetrahydrocannabivarin (THCV), cannabichromene (CBC), cannabigerol (CBG), cannabinol (CBN), Δ9-tetrahydrocannabinol (THC)) in pancreatic beta cell differentiation via the WNT5A/JNK and BMP pathways using an in-silico approach.
Specifically, we aim to elucidate the molecular mechanisms by which cannabinoids modulate these pathways by targeting key proteins involved in these pathways: BMP6, SMAD 1/4/5 (for BMP pathway), and WNT5A, JNK, c-JUN, and FZD3 (for WNT5A/ JNK pathway). Our innovative approach utilizes computational methods to predict the binding affinity and molecular interactions of cannabinoids with key proteins involved in these pathways, providing a novel and efficient strategy for identifying potential therapeutic targets for diabetes treatment. By gaining a deeper understanding of the molecular mechanisms underlying the effects of cannabinoids on beta cell differentiation and function, we can pave the way for the development of new and effective therapies for diabetes.
Methodology
Optimization of Protein and Ligand
The ligands (CNG: CID 5,315,659; CBC: CID 30,219; THCV: CID 93,147; CBN: CID 2543; CBD: CID 644,019; THC: CID 16,078) were downloaded as SDF files from PubChem and transformed into pdbqt files using the open Babel tool of PyRx (Dallakyan, 2010). The target protein structures of SMAD4 (PDB ID; 1u7v), SMAD1 (PDB ID; 1khu), cJUN (PDB ID; 4YR8), SMAD5 (PDB ID; 6fzs), FDZ3 (PDB ID; 7x8t), BMP6 (PDB ID; 2r53) were retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/) [13]. The target proteins were prepared by removing water molecules and adding polar hydrogens and charges using AutoDock [14]. Chimaera was used to visualise and modify the target molecule and remove the ligand. The protein was also visualised using Biovia Discovery Studio Visualizer version 24.1.0.23298 (BIOVIA 2023).
Virtual Screening of Molecules by Pyrx Tool Docking simulation and Analysis
The docking was performed by using PyRx (Autodock vina) tools version v0.8 programs [15]. Atomic solvation parameters were added and Kollman charges were assigned to target proteins. The internal degrees of freedom and torsions were set, the non-polar hydrogen was combined with the carbons, and polar hydrogen charges of the Gasteiger type were assigned. The target protein complex was docked with cannabinoid compounds, where the ligand was flexible and the protein was considered as a rigid body. After loading the protein into the PyRx program, a PDBQT file containing the protein structure with hydrogens in each polar residue was created. Every ligand bond was configured to be rotatable. For protein-fixed ligand-flexible docking, the Lamarckian Genetic Algorithm (LGA) approach was utilized for all calculations. The various complexes were sorted according to the anticipated binding energy, and the findings were evaluated. A cluster analysis was carried out using root mean square deviation values concerning the initial geometry, and the populated cluster with the lowest energy conformation was considered.
Validation of Protein-Ligand Interactions
The PyRx Virtual screening tool was performed to validate the protein-ligand complex by rmsd/ub and rmsd/lb values. The mean distance between atoms of a position concerning the best fitting position is measured using only movable heavy atoms to compute the Root Mean Square Deviation (RMSD) values. Two distinct RMSD measures were given: rmsd/lb (the RMSD lower bound) and rmsd/ ub (the RMSD upper bound). The RMSD metrics rmsd/ub (RMSD upper bound) and rmsd/lb (RMSD lower bound) are provided as alternatives. Rmsd/ub compares each atom in one conformation to itself in the other conformation.
Molecular Dynamics Simulation of the Ligand-Receptor Complex
The iMOD server was used to execute molecular dynamics simulations. The stability and flexibility of protein and ligand complex was assessed by iMODS server (http://imods.Chaconlab. org/) which carries out Normal Mode Analysis (NMA) in internal coordinates (dihedral) using an elastic network model [16]. The server illustrates the stability of the complex by providing deformability plots, covariance map, eigenvalues, and elastic network.
Results
Outcomes of Molecular Docking Analysis
The molecular docking results provided in Table 1 give insights into the binding interactions between six cannabinoids (THC, CBN, CBC, CBG, CBD, THCV) and proteins involved in the BMP Pathway (BMP6 and SMAD1/4/5) and WNT5A/JNK (WNT5A, JNK, c JUN, and FZD3). All of the cannabinoids show potential to interact with the target proteins, indicated by binding affinities ranging from -4.7 to -8.6 kcal/mol. More negative values signify stronger binding. The highest affinity was observed for the JNK-CBN complex (-8.6 kcal/ mol). THCV also displayed strong binding to SMAD4 (-8.4 kcal/ mol). In general, CBN and THCV tended to show maximum binding scores compared to the other cannabinoids.
Table 1:Biding affinities of cannabinoids with proteins related to Pancreatic Beta Cell Differentiation.
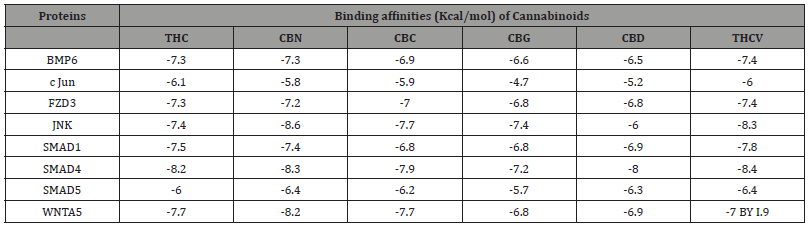
CBC: Cannabichromene; CBD: Cannabidiol; CBG: Cannabigerol; CBN: Cannabinol; THC: Δ9-tetrahydrocannabino; l THCV: Tetrahydrocannabivarin; BMP: Bone Morphogenetic Protein; c-Jun: Cellular Jun proto-oncogene; FZD3: Frizzled Class Receptor 3; JNK: c-Jun N-terminal Kinase; SMAD: Mothers Against Decapentaplegic Homolog; WNTA5: Wingless-Type MMTV Integration Site Family, Member 5A.
Protein-ligand complex Validation by RMSD/lb and RMSD/ub
Table 2 reports the Root Mean Square Deviation (RMSD) values calculated for the protein-ligand complexes obtained from molecular docking. The RMSD values validate the accuracy and stability of the binding complexes. RMSD/lb (lower bound) and RMSD/ub (upper bound) values indicate the average distance deviations between atoms in the docked conformations compared to the original or best-fitted positions. Acceptable RMSD/lb values are generally below 2.0 Å, suggesting minimal structural changes after binding. The RMSD/lb values for all complexes in this study fell between 1.203 to 2.022 Å, well within the acceptable range. This implies that docking was successful in identifying stable, low-energy binding modes for the cannabinoid ligands within the target protein pockets. Similarly, the RMSD/ub values ranging from 3.677 to 10.038 Å demonstrate that the docked structures closely matched the original protein-ligand conformations. Overall, the low RMSD scores validate that binding did not distort or damage the protein-ligand structures.
Table 2:Root mean square deviation (RMSD) of values lower and upper bounds.
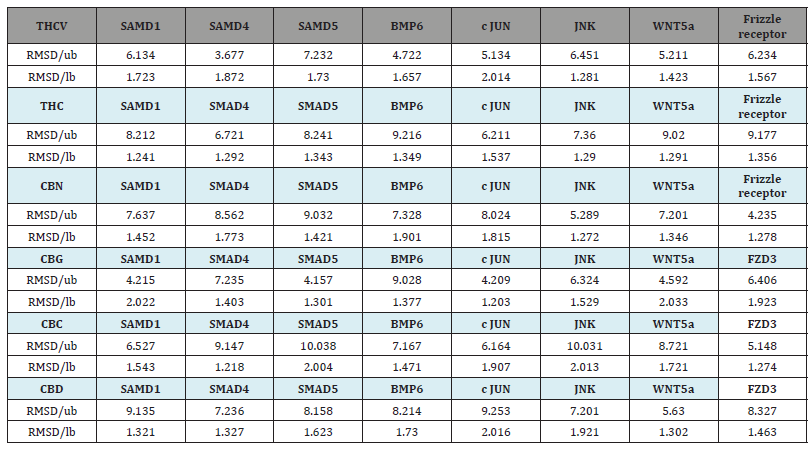
CBC: Cannabichromene; CBD: Cannabidiol; CBG: Cannabigerol; CBN: Cannabinol; THC: Δ9-tetrahydrocannabino; l THCV: Tetrahydrocannabivarin; BMP: Bone Morphogenetic Protein; c-Jun: Cellular Jun proto-oncogene; FZD3: Frizzled Class Receptor 3; JNK: c-Jun N-terminal Kinase; SMAD: Mothers Against Decapentaplegic Homolog; WNTA5: Wingless-Type MMTV Integration Site Family, Member 5A; RMSD/ub: Root ean Square Deviation; ub: upper bound; lb: lower bound.
Outcomes of 3D visualization of the main binding interactions
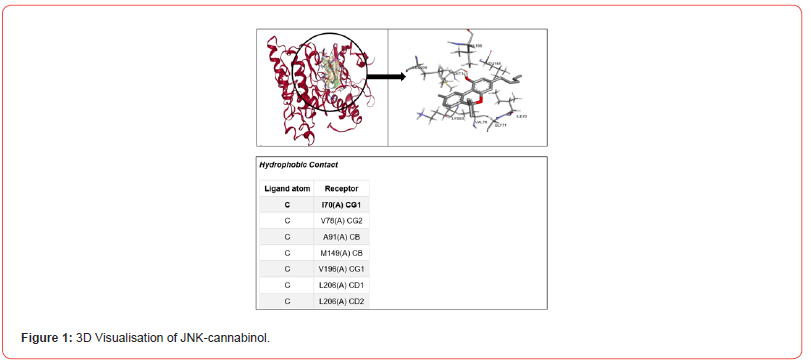
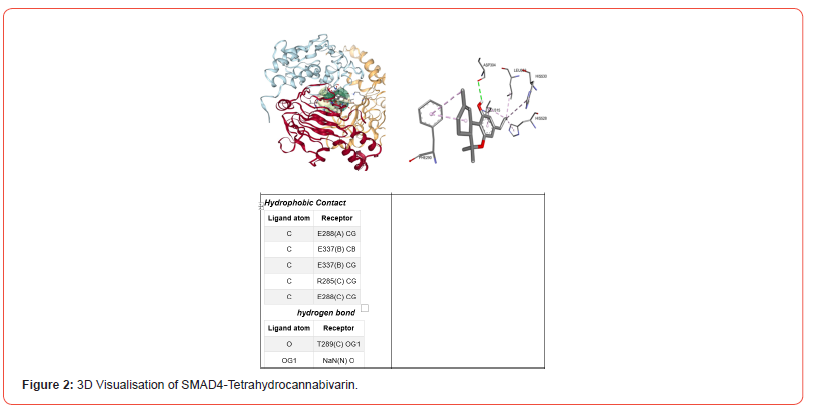
Figure 1 shows the 3D visualization of the binding interactions between Cannabinol and JNK. It indicates that Cannabinol interacts with JNK through hydrophobic contacts, forming alkyl bonds between the ligand carbons and receptor amino acid carbons (I70, V78, A91, M149, V196, L206). These non-covalent hydrophobic bonds stabilize the ligand orientation within the protein binding pocket. Figure 2 depicts the binding mode of tetrahydrocannabivarin with SMAD4. It suggests tetrahydrocannabivarin engages in both hydrophobic contacts and a hydrogen bond with SMAD4. Hydrophobic interactions occur between ligand and receptor carbons (E288, E337, R285, E288). Additionally, there is a hydrogen bond between the ligand oxygen and receptor oxygen (O-T289).
Results of Molecular Dynamics Simulation
Figure 3 shows the outputs of normal mode analysis (NMA) conducted on the SMAD4-THCV and JNK-CBN protein-ligand complexes obtained from molecular docking. NMA provides insights into the structural flexibility and deformation of biomolecules. Figure 3a displays the deformation plot, which visualizes the displacements along low frequency normal modes. This helps identify flexible and rigid regions within the complexes. Figure 3b shows the eigenvalue spectrum. Lower eigenvalues correspond to softer, more deformable motions. The distinct pattern of eigenvalue peaks and valleys indicates variation in complex stiffness across different atomic motions. Figure 3c presents the covariance map, where differing color tones represent correlated (red), uncorrelated (white) or anticorrelated (blue) atomic motions within the complexes. This provides information on cooperative structural changes. Figure 3d depicts the elastic network, where darker gray dots signify stiffer springs between atom pairs in the bonded network model.
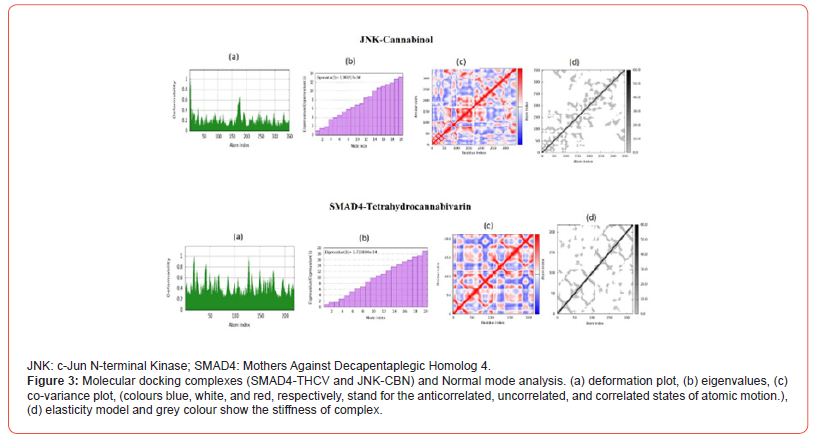
Discussion
The WNT5A/JNK and BMP pathways play crucial roles in pancreatic beta cell differentiation, proliferation, and function, and their dysregulation has been implicated in the pathogenesis of diabetes [5]. Targeting the WNT5A/JNK and BMP pathways with small molecules or biologics could modulate beta cell differentiation, proliferation, and function, leading to improved glycemic control and reduced diabetes-related complications. The aim of this study was to investigate the role of cannabinoids from Cannabis sativa in pancreatic beta cell differentiation via WNT5A/ JNK and BMP pathways. A total of 8 proteins were targeted, BMP6 and SMAD 1/4/5 for BMP Pathway, and WNT5A, JNK, c JUN, and FZD3 for WNT5A/JNK pathway. All the six cannabinoids (THC, CBN, CBC, CBG, CBD, THCV) interacted with targets in both pathways, suggesting their potential to modulate beta cell differentiation through these molecular mechanisms (Table 1). To validate the stability and accuracy of the protein-cannabinoid complexes obtained from molecular docking simulations, RMSD values (Table 2).
The target proteins analyzed (SMAD1/4/5, BMP6, WNT5A, JNK, c-JUN, and FZD3), are involved at different levels in the signaling cascades that regulate beta cell fate and function. The RMSD results confirm that docking yielded stable, low-energy binding conformations for cannabinoids within target protein pockets, suggesting that cannabinoids could directly interfere with native protein-protein interactions by competitive inhibition. The findings of the present study demonstrate the precision of the docking methodology used, as evidenced by the rmsd/lb values being below the established threshold of 2.0 for assessing reliability [17]. This indicates that the docking approach employed in this investigation is accurate and reliable. The Table 1 shows that the main interactions with noted between CBN and JNK in the WNT5A/JNK pathway, and between THC and SMAD4 in the BMP pathway. The c-Jun N-terminal kinase (JNK) is a key component of the WNT5A/ JNK pathway. Activation of JNK has been shown to promote beta cell apoptosis and dysfunction, while its inhibition leads to beta cell proliferation and survival [18,19].
SMAD4 is a central mediator of the BMP pathway, which is essential for beta cell proliferation, survival, and function. SMAD4 acts as a signal transducer in the BMP pathway, forming complexes with phosphorylated SMAD proteins that translocate to the nucleus to regulate gene transcription [20,21]. Targeting JNK and SMAD4 signaling pathways has emerged as a promising strategy for the development of novel anti-diabetic therapies. For example, small molecule inhibitors of JNK have been shown to improve beta cell function and glucose homeostasis in diabetes [22]. Similarly, modulating BMP signaling through the use of BMP agonists or antagonists has been shown to enhance beta cell proliferation and function, and improve glycemic control in animal models of diabetes [23]. Understanding the interactions between proteins and ligands is essential for developing computational models of molecular recognition and functional inhibition.
These interactions can be classified into four primary types: ionic, hydrophobic, hydrogen bonding, and water bridges [24]. The binding interactions using 3D visualization techniques was performed between CBN-JNK and THC-SMAD4 (Figures 1&2). CBN interacts with JNK residues via hydrophobic contacts and THC interact with SMAD4 residues via hydrophobic contacts and a hydrogen bond. It was reported that hydrogen bonding, plays a vital role in the proper folding of proteins and their interactions with ligands. These bonds are essential for maintaining the three-dimensional structure of proteins and stabilizing proteinligand complexes [25]. On the other hand hydrophobic bonds helps to stabilize the three-dimensional structure of proteins and contributes to the formation of protein-ligand complexes by promoting the binding of nonpolar ligands to hydrophobic pockets on the protein surface [26]. Validating these ligand poses enhances confidence that CBN and THC may directly modify key proteins governing beta cell fate.
To gain insights into molecular flexibility and deformation, normal mode analysis (NMA) was conducted on the SMAD4-THCV and JNK-CBN protein-ligand complexes from molecular docking. Figure 3 depicts the outputs of the NMA, revealing flexible and rigid regions within the complexes that could impact interaction surfaces, variation in complex stiffness across motions, correlated, uncorrelated, and anticorrelated atomic motions within the complexes, and ligand-induced changes to intrinsic protein elasticity [27]. The NMA results validate that docked complexes retained structural integrity while providing new insights into cannabinoid-induced modulation of protein dynamics.
Conclusion
This study showed that all six cannabinoids (THC, CBN, CBC, CBG, CBD, THCV) had binding affinities with proteins involved in WNT5A/JNK and BMP pathways, with the highest affinity observed for the JNK-CBN and SMAD4-THCV. The results also validated the accuracy and stability of the protein-ligand complexes obtained from molecular docking simulations, while normal mode analysis offered additional insights into molecular flexibility and deformation. While further in vitro and in vivo studies are necessary to validate these findings, this in silico approach offers a valuable starting point for exploring the therapeutic potential of cannabinoids in diabetes treatment by targeting beta cell differentiation pathways.
Authors’ Contributions
GRT designed the study; GRT and SI: wrote the manuscript; SI performed in silico analysis; FAE, CFT and CRN reviewed and revised the manuscript; and MGM supervised the study and the manuscript writing. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no financial or non-financial interests that are directly or indirectly related to the work.
Acknowledgments
We acknowledge the researchers of the IK-based Technology Innovations at the Department of Science and Innovation, University of the Free State, Bloemfontein, South Africa.
References
- Izzo AA, Capasso R, Aviello G, Borrelli F, Romano B, et al. (2012) Inhibitory effect of cannabichromene, a major non-psychotropic cannabinoid extracted from Cannabis sativa, on inflammation-induced hypermotility in mice. Br J Pharmacol 166(4): 1444-1460.
- Rorsman P, Ashcroft FM (2018) Pancreatic β-Cell Electrical Activity and Insulin Secretion: of Mice and Men. Physiol Rev 98(1): 117-214.
- Ram Makena M, Gatla H, Verlekar D, Sukhavasi S, Pandey M, et al. (2019) Wnt/β-Catenin Signaling: The Culprit in Pancreatic Carcinogenesis and Therapeutic Resistance. Int J Mol Sci 20(17): 4242.
- Wang RN, Green J, Wang Z, Deng Y, Qiao M, et al (2014) Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 1(1): 87-105.
- Chmielowiec J, Szlachcic WJ, Yang D, Scavuzzo MA, Wamble K, et al. (2022) Human pancreatic microenvironment promotes β-cell differentiation via non-canonical WNT5A/JNK and BMP signaling. Nat Commun 13(1): 1952.
- Yung T, Poon F, Liang M, Coquenlorge S, McGaugh EC, et al. (2019) Sufu- and Spop-mediated downregulation of Hedgehog signaling promotes beta cell differentiation through organ-specific niche signals. Nat Commun 10(1): 4647.
- Liu Y, Du SY, Ding M, Dou X, Zhang FF, et al. (2017) The BMP4-Smad signaling pathway regulates hyperandrogenism development in a female mouse model. J Biol Chem 292(28): 11740-11750.
- Chenglin Wang, Yuan Zhao, Yingying Su, Ruimin Li, Yunfeng Lin, et al. (2013) C-Jun N-Terminal Kinase (JNK) Mediates Wnt5a-Induced Cell Motility Dependent or Independent of RhoA Pathway in Human Dental Papilla Cells. PLoS One 8(7): e69440.
- González Mariscal I, Pozo Morales M, Romero Zerbo SY, Espinosa Jimenez V, Escamilla Sánchez A, et al. (2022) Abnormal cannabidiol ameliorates inflammation preserving pancreatic beta cells in mouse models of experimental type 1 diabetes and beta cell damage. Biomed Pharmacother 145: 112361.
- Jourdan T, Godlewski G, Kunos G (2016) Endocannabinoid regulation of β-cell functions: implications for glycaemic control and diabetes. Diabetes Obes Metab 18(6): 549-557.
- Ghasemi Gojani E, Kovalchuk I, Kovalchuk O (2022) Cannabinoids and terpenes for diabetes mellitus and its complications: from mechanisms to new therapies. Trends Endocrinol Metab 33(12): 828-849.
- Yujeong Kim, Wonhee Kim, Soo Hyun Kim, Kyu Sang Sim, Ki Hyun Kim, et al. (2023) Protective Effects of Hemp (Cannabis sativa) Root Extracts against Insulin-Deficient Diabetes Mellitus in Mice. Molecules 28(9): 3814.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28(1): 235-242.
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31(2): 455-461.
- Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263: 243-250.
- Benhander GM, Abdusalam AAA (2022) Identification of Potential Inhibitors of SARS-CoV-2 Main Protease from Allium roseum L. Molecular Docking Study. Chemistry Africa 5(1): 57-67.
- López Blanco JR, Aliaga JI, Quintana Ortí ES, Chacón P (2014) iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 42: W271-276.
- Solinas G, Becattini B (2017) JNK at the crossroad of obesity, insulin resistance, and cell stress response. Mol Metab 6(2): 174-184.
- Yung JHM, Giacca A (2020) Role of c-Jun N-terminal Kinase (JNK) in Obesity and Type 2 Diabetes. Cells 9(3): 706.
- Chi LH, Redfern AD, Roslan S, Street IP, Burrows AD, et al. (2024) Loss of tumor-derived SMAD4 enhances primary tumor growth but not metastasis following BMP4 Cell Commun Signal 22(1): 248.
- Perera N, Ritchie RH, Tate M (2020) The Role of Bone Morphogenetic Proteins in Diabetic Complications. ACS Pharmacol. Transl Sci 3(1): 11-20.
- Vernia S, Cavanagh Kyros J, Barrett T, Tournier C, Davis RJ (2016) Fibroblast growth factor 21 mediates glycemic regulation by hepatic JNK. Cell Rep 14(10): 2273-2280.
- Bruun C, Christensen GL, Jacobsen MLB, Kanstrup MB, Jensen PR, et al. (2014) Inhibition of beta cell growth and function by bone morphogenetic proteins. Diabetologia 57(12): 2546-2554.
- Rashid HU, Ahmad N, Abdalla M, Khan K, Martines MAU, et al. (2022) Molecular docking and dynamic simulations of Cefixime, Etoposide and Nebrodenside A against the pathogenic proteins of SARS-CoV-2. J Mol Struct 1247: 131296.
- Yunta Maria JR (2017) It Is Important to Compute Intramolecular Hydrogen Bonding in Drug Design. American Journal of Modeling and Optimization 5(51): 24-57.
- Ferenczy GG, Kellermayer M (2022) Contribution of hydrophobic interactions to protein mechanical stability. Comput Struct Biotechnol J 20: 1946-1956.
- Bauer JA, Bauerová Hlinková V (2020) Normal Mode Analysis: A Tool for Better Understanding Protein Flexibility and Dynamics with Application to Homology Models. In book: Homology Molecular Modeling- Perspectives and Applications.
-
Guy Roussel Takuissu*, Shabnor Iqbal, Cedric Fossi Tchinda, Fils Armand Ella, Corinne Raissa Ngnameko, Matsabisa Gilbert Motlalepula*. Investigating the Role of Cannabinoids from Cannabis sativa in Pancreatic Beta Cell Differentiation via WNT5A/JNK and BMP Pathways: An In-Silico Approach. Curr Tr Clin & Med Sci. 4(1): 2025. CTCMS.MS.ID.000578.
-
epidemiology, Enterobacteriaceae, infection pneumonia, urinary tract infection
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






