Research Article
Research Article
NPHS1 and NPHS2 Variants Associated with Early-Onset Frequently Relapsing or Steroid-Dependent Nephrotic Syndrome
Ahmad O Babalghith1, Nasser A Elhawary1*, Iman S Abumansour1, Ehab M Melibary1, Ghydda Alghamdi1, Ikhlas A Sindi2, Ezzeldin N Elhawary3, Najiah M Alyamani4, Ibtesam S Almami5, Hind Naffadi6, Afnan Shakoori7, Luai Zamil8, Nasser Alenezy9 and Mohammed Dandini10
1Department of Medical Genetics, College of Medicine, Umm Al-Qura University, Mecca, SA
2Department of Biotechnology, Faculty of Science, King Abdulaziz University, SA
3MS Genomic Medicine Program, Faculty of Medicine, University of Southampton, Southampton General Hospital, UK
4Department of Biology, College of Science, University of Jeddah, Jeddah, SA
5Department of Biology, College of Science, Qassim University, SA
6Common Science, First Year Deanship, Umm Al-Qura University, Mecca, SA
7Department of Laboratory Medicine, Faculty of Applied Medical Sciences, Umm Al-Qura University, Mecca, SA
8Department of Laboratory and Blood Bank, King Abdulaziz Hospital, Mecca, SA
9Rumah General Hospital, Ministry of Health; Riyadh, SA
10Department of Laboratory and Blood Bank, Maternity and Children Hospital, Mecca, SA
Nasser A Elhawary, Department of Medical Genetics, Umm Al-Qura University, Mecca 21955, P.O. 57543, Saudi Arabia.
Received Date: June 17, 2022; Published Date: June 29, 2022
Abstract
Background: Glomerular podocytes and slit diaphragms remain the major causes of progressive proteinuria and nephrotic syndrome (NS).
Objective: Here, we analyzed genetic variants in the nephrin (NPHS1) and podocin (NPHS2) genes in cases with early-onset presentation of NS in Saudi Arabia.
Subjects and methods: Clinical characteristics and genomic DNA were collected from 35 unrelated NS cases (mean age of onset, 3.0 years; range, stillbirth-8 years). Cases were diagnosed based on histologic kidney biopsy and biochemical parameters. Enzymatic digestion was performed on PCR amplicons for the frameshift L41NfsX50 and nonsense R1109X variants of NPHS1, and the entire coding regions of NPHS2 were sequenced.
Results: We identified 15 cases of frequently relapsing NS or steroid-dependent NS (FRNS/SDNS) (42.9%), 12 cases of steroid-resistant NS (SRNS) (34.3%), and 8 cases of steroid-sensitive NS (SSNS) (22.9%). Cases with FRNS/SDNS (46.2%) had a younger age at presentation than cases with SSNS (38.5%; P = 0.52) or SRNS (15.4%; P = 0.0056). one SSNS case, three FRNS/SDNS, and three SRNS carried the NPHS1 R1109X (Finminor) variant. For NPHS2, one SSNS case carried a homozygote R168H variant, and another SSNS case carried a compound heterozygote R229Q variant. The allele frequencies of the NPHS1 R1109X variant and the NPHS2 R168H and R229Q variants were 14% and 4.3%, respectively. No cases carried the NPHS1 L41NfsX50 variant. Available kidney biopsy reports revealed that minimal change disease was most frequent in FRNS/SDNS cases (56.7%) and focal segmental glomerulosclerosis, most common in SRNS cases (75%).
Conclusion: A significant proportion of the youngest SSNS cases may develop FRNS/SDNS.
Keywords: Nephrotic syndrome; NPHS1; NPHS2; Nephrin; Podocin; Frequently-relapsing or Steroid-dependent; Steroid-resistance
Abbreviations: CRF -chronic kidney failure; FATHMM-Functional Analysis through Hidden Markov Models; FRNS -Frequently Relapsing Nephrotic Syndrome; FSGS- Focal Segmental Glomerulosclerosis; GFR- Glomerular Filtration Rate; IgA- Immunoglobulin A; ISKDC-International Study of Kidney Disease in Children; Lo Ftool- loss-of-Function Tool; MCD-Minimal Change Disease; MCNS-Minimal change Nephrotic Syndrome; MsPGNMesangioproliferative glomerulonephritis; NPHS1-Nephrin; NPHS2-podocin; NS- Nephrotic Syndrome; PolyPhen-2- Polymorphism Phenotyping v2, SA- Saudi Arabia SDNS- Steroid-dependent Nephrotic Syndrome; SIFT- Sorting Intolerant from Tolerant, SRNS -steroid-Resistant Nephrotic Syndrome; SSNS- Steroid-Sensitive Nephrotic Syndrome
Introduction
Nephrotic syndrome (NS) is the most common diagnosis in pediatric nephrology and is evolving as a leading cause of uremia [1]. Affected individuals have massive proteinuria in utero, and NS develops soon after birth (especially in premature infants) [2]. Associated with proteinuria, other visible signs that appear soon after birth include hypoalbuminemia, hyperlipidemia, edema, and abdominal distention [2]. Electron microscopy of the kidneys in patients with NS has shown blurring of the podocytes and the absence of split diaphragms. Early bilateral nephrectomy followed by kidney transplantation in infants is the only curative therapy for NS, and the disorder is otherwise lethal [3 ]. Thus, NS diagnoses are based on the clinical features of the affected neonate and kidney histopathology. NS can be suspected prenatally based on increased α-fetoprotein in the maternal serum or amniotic fluid [4].
For decades, NS has been categorized, based on its clinical response to steroid medications, as either steroid-sensitive NS (SSNS) or steroid-resistant NS (SRNS) [5]. About 80% of NS cases are steroid sensitive. Of these individuals with SSNS, 70% will suffer from frequent relapses, and a significant proportion may develop frequently relapsing NS or steroid-dependent NS (FRNS/ SDNS) [1]. Children with FRNS/SDNS are often treated with repeated corticosteroid courses [6], as a second-line treatment, with non-corticosteroid immunosuppressive medications, often associated with heavy treatment burden [7]. In addition, 20% of sporadic children with SRNS may face a therapeutic obstacle to pediatricians and nephrologists. Kidney histology revealed focal segmental glomerulosclerosis (FSGS), minimal change nephrotic syndrome (MCNS), and mesangioproliferative glomerulonephritis (MsPGN) in 75%, 20% and 5% of the SRNS patients, respectively [8-10]. In about 14% of patients with FSGS [11,12], deformities in the podocytes and split diaphragm can be explained by genes such as INF2 (MIM 610982), ACTN4 (MIM604638), TRPC6 (MIM 603652), WT1 (MIM 607102), NPHS1 (MIM 602716), and NPHS2 (MIM 604766) [11]. Treatment with cyclosporin A is much more effective in patients with nonhereditary SRNS than in those with geneticbased SRNS [12]. suggesting that genetic variation contributes to the pathogenesis of FSGS and influences the outcome of clinical treatment. Thus, it is of great clinical significance to elucidate the genetic characteristics of FSGS.
Variants in the NPHS1 gene have been identified as a cause of congenital NS of the Finnish type [13], and NPHS1 is one of the most common genes associated with SSNS. NPHS1 is located at chromosome 19q13.1 and contains 29 exons spanning 150 kb. Nephrin, the transmembrane protein encoded by NPHS1, is produced only in glomerular podocytes, where it participates in the intercellular junctions of mature podocytes and slit diaphragm formation [14]. Nephrin also serves as the main component of the glomerular filtration barrier [15]. Reduced expression of nephrin could result in progressive proteinuria with glomerular hypertrophy and FSGS [16]. The prevalence of NPHS1-associated NS has increased worldwide, although more than half of the cases were diagnosed in Finland, where the prevalence is 1 in 8,200 births [17]. Despite its association with congenital NS of the Finnish type, NPHS1 has been confirmed as a susceptibility gene for various kidney diseases, including SRNS, FSGS, MCD, and IgA nephropathy [14,18-21].
Among Finnish children with congenital NS of genetic origin, nearly 78% have the p.Leu41fsX90 variant in exon 2 of NPHS1, and nearly 16% have the p.R1109X variant in exon 26 of NPHS1 [13]; these variants, which are referred to as Fin-major and Fin-minor, respectively [22,23], have rarely been reported in non-Finnish ethnic populations [14]. NPHS1 genetic screening of non-Finnish patients has shown that the frequency of NPHS1 variants is lower than that in Finnish patients (39% versus 55% of NS cases) [18,20].
Most patients with idiopathic NS respond to steroid therapy and show a favorable outcome, but 20% are resistant to steroid therapy, with progression to end-stage kidney failure in many cases. A subgroup of nephrosis referred to as steroid-resistant idiopathic NS has been investigated [22] and is characterized by familial occurrence, age of onset in early childhood, resistance to steroid therapy, progression to end-stage kidney disease within a few years, and absence of recurrence after kidney transplantation.
NPHS2 is a gene encoding podocin, another important protein located at the split diaphragm. Podocin is an integral protein consisting of 383 amino acids having a single short hairpin-like transmembrane domain and cytosolic N- and C-terminal domains [24]. It is expressed in the podocyte-foot process cell membrane at the split diaphragm’s insertion site and might maintain the integrity of the split diaphragm [25]. Pathogenic variants in NPHS2 can result in a reduction or absence of functional proteins and thus impair the formation of normal slit diaphragms [26,27]. This abnormality leads to SRNS before six years of age, and patients reach end-stage kidney disease during their first decade of life [26]. Several reports have demonstrated that variants in the NPHS2 gene represent a frequent cause of SRNS, occurring in 20-30% of sporadic SRNS cases [27,28], NPHS2 gene is frequently associated with FSGS, where their variants in NPHS2 can cause autosomal recessive SRNS. Several genetic variants of the NPHS2 gene, including R229Q and R291W, have been described in patients with NS, and individuals carrying both variants have shown progressive loss of kidney function. The p.R229Q variant (c.686G>A; rs61747728), a nonsynonymous variant in exon 5, is affirmed to be one of the most important predictive factors in the pathogenesis of SRNS [29-31]. Evidence has confirmed that the R229Q allele is a disease-causing variant rather than a benign polymorphism [29]. However, susceptibility in individuals with R229Q variants varies among ethnic groups. A wide range of incidence in pediatric idiopathic NS syndrome worldwide is reported to be 1.15-11.6 per 100,000 children [32], whereas the risk of average annual incidence has been reported higher in Asian (7.14/100,000) and Black (3.53/100,000) children compared to White Caucasian (1.83/100,000), and or American (2.13/100,000) children.
Given the NPHS1 and NPHS2 variants commonly reported worldwide in recent decades, the present study analyzed these variants in Saudi children with the early-onset presentation of NS and identified the types of NS and phenotypes associated with the different variants.
Subjects and Methods
Ethics statement and consent
Molecular analyses were performed at the DNA Diagnostic Laboratories, Medical Genetics Department, College of Medicine, Umm Al-Qura University. This study was conducted under the Declaration of Helsinki. The Institutional Biomedical Ethics Committee-Umm Al-Qura University (reference #HAPO-02-K-012), licensed from the National Committee of Medical and Bioethics (King Abdelaziz City for Science and Technology-Riyadh), approved the study. The parents or guardians of 35 children with NS and 40 healthy controls gave informed consent before participating in the study.
Definition and management
We followed the protocol of the International Study of Kidney Disease in Children (ISKDC) [33] ,which categorized NS cases based on their response to treatment: 1) Uncomplicated SSNS: relapse ≤ 4 times/year and have not required treatment with steroid-sparing agents. 2) SRNS: no response to steroid treatment after four weeks of enteral prednisone at 60 mg/m2 per day in addition to three intravenous administrations of methylprednisolone at 1,000 mg/1.73 m2 per dose. 3) FRNS/SDNS: relapse ≥ 2/month (FRNS) or have had two consecutive relapses during the tapering period or relapsed within two weeks of discontinuing steroid therapy (SDNS).
The Schwartz formula was used to estimate the glomerular filtration rate (GFR) in NS cases. However, GFR = k * height (cm)/ plasma creatinine (mg/dL): k is 0.45 for infants (age ≤ 1.5 years), 0.55 for older children and adolescent girls, and 0.70 for adolescent boys > 13 years of age. Decreased kidney function was defined as GFR < 90 ml/min/1.73m2, chronic kidney failure (CRF) as GFR < 60 ml/min/1.73m2, and end-stage kidney disease as GFR < 15 ml/ min/1.73m29 [5].
Study population
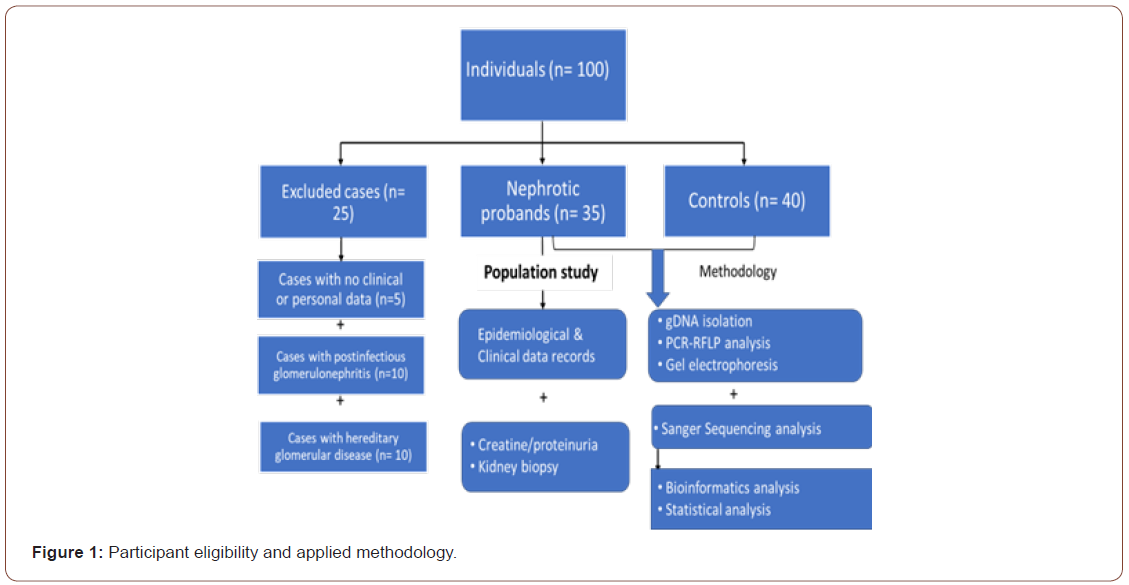
A total of 35 children with histologically proven NS were included in this study. Cases were diagnosed in pediatric nephrology clinics of different hospitals by nephrologists in Western regions of Saudi Arabia, including Makkah, Al-Hada, and Al-Taif governorates. The diagnosis of NS was established based on histological examination of a specimen of kidney tissue gained by kidney biopsy. The clinical diagnosis of NS required the presence of heavy proteinuria (>50 mg/kg/day or urine protein/creatinine ratio ≥ 2 mg/mg) and hypoalbuminemia (<2.5 mg/dl). Clinical records were gathered, including family history, age at onset, gender, frequency of relapse, kidney biopsy reports, GFR, and treatment modes. Cases with postinfectious glomerulonephritis and systemic diseases (e.g., lupus erythematosus, diabetes, amyloidosis, vasculitis, metabolic or toxic nephritis, hepatitis B, or hereditary glomerular disease) were excluded from this study. Controls included 40 children with no personal or family history of kidney disease (Figure 1).
DNA analysis
Genomic DNA samples were isolated from buccal cells by gently scraping the mucosa using an Oragene DNA-OGR-575 kit (DNA Genotek Inc, Ottawa-ON, Canada) following some modifications [34 ]. Sometimes, DNA was isolated from peripheral blood samples using a DNA extraction Spin Column kit (Qiagen, USA).
PCR Conditions
Genomic DNA was subjected to a 25-μl reaction volume of 0.5 mM of each primer, 200 μM of each dNTP, 67 mM Tris-HCl, 16 mM (NH4)2SO4, 0.01% Tween-20, 1 mM MgCl2, and 0.15 units of Taq DNA polymerase. The samples were then subjected to DNA denaturation at 95°C for 5 min, followed by 30 cycles of denaturing for 1 min at 95°C, annealing at 58°C for 30 s, and extension for 30 s at 72°C, with a final extension of 7 min at 72°C in a PCR Engine Dyad thermocycler (Bio-Rad Laboratories Inc, Hercules, CA). The primers were designed based on previously published sequences for the NPHS1 gene (L41NfsX50 and R1109X loci) and the NPHS2 gene (R229Q, and R291W loci) [13,35 ]. Oligonucleotide primer sequences are listed in [Table 1].
Table 1: Amplification conditions including oligonucleotide sequences, enzymatic temperature, and fragment sizes.

Enzymatic digestion of NPHS1 and NPHS2 Loci
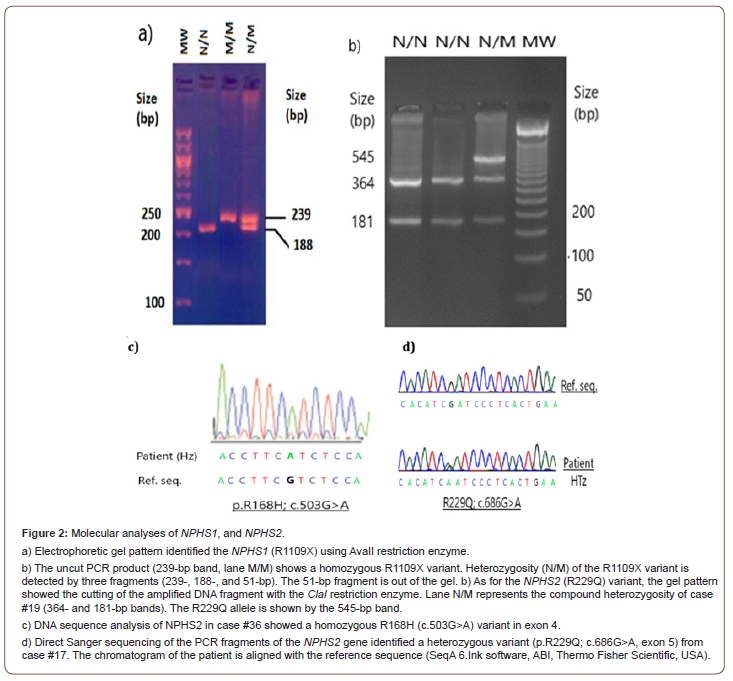
We identified the NPHS1 (L41Nfs50 and R1109X) and NPHS2 (R229Q and R291W) variants using enzymatic digestion of their PCR fragments. Briefly, the PCR amplicons of these variants were incubated with TaqI, AvaII, ClaI, and PflMI endonucleases for 2 hours according to the manufacturer’s instructions (New England Biolabs Inc., MA, USA) (Table 1). The digested fragments were separated on a 3%MetaPhor agarose gel (Lonza Rockland, Inc., USA) for 2 hours at 3.5 volts/cm. The digested fragments were visualized under a UV transilluminator and photographed using a Gel Documentation System (UVitec, Cambridge, UK). Genotyping results were confirmed by sequencing 20% of the samples (Figure 2a,b).
Sequence analysis of the whole NPHS2 Gene
For mutation analysis, Sanger sequencing was carried out using oligonucleotide primers optimized for all exonic regions of NPHS2. Briefly, PCR amplicons were purified using SPRI paramagnetic bead-based chemistry to remove contaminants, including dNTPs, salts, primers, and primer dimers, according to the manufacturer’s instructions (AMPure XP, Beckman Coulter Life Sciences, IN, USA). The purified products were treated with the Big Dye Terminator v3.1 cycle sequencing kit and then sequenced using the 3500 Genetic Analyzer sequencer system (Thermo Fisher Scientific, USA). Oligonucleotide sequences of eight exons, including flanking regions, were previously reported (Table S1). The full oligonucleotide primers optimized for NPHS2 gene can be found in the Supplement (see supplementary file). Variants were reviewed and aligned using dbSNP (Sequence Analysis Software ver. 6, ABI, Thermo Fisher, USA) (Figure 2c,d).
Bioinformatics analysis
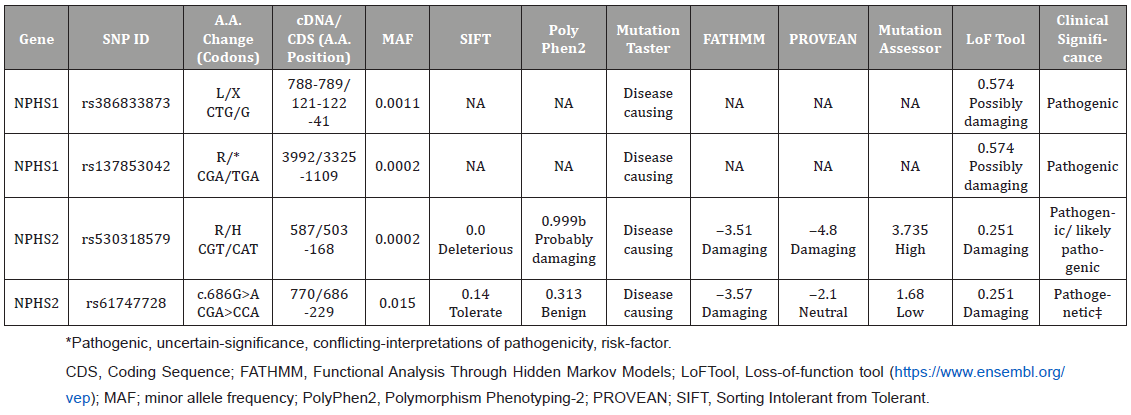
For nonsynonymous variants, we used the in silico tools Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen2), MutationTaster, Functional Analysis through Hidden Markov Models (FATHMM), Mutation Assessor, and lossof- function tool (LoFtool) to predict the effects of the variants on their corresponding functional proteins (https://www.ensembl.org/vep/) (Table 2).
Table S1: Oligonucleotide primers optimized for NPHS2 gene.
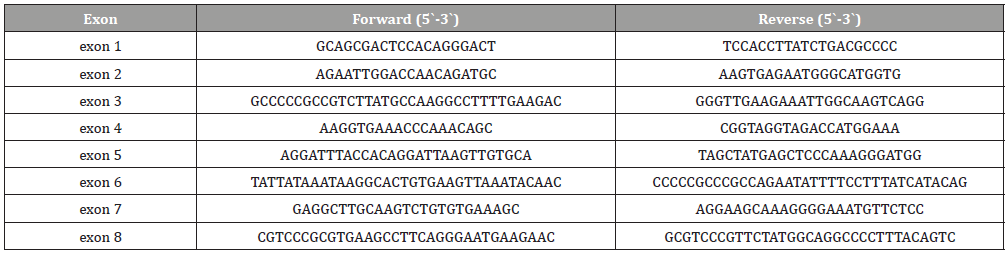
Table 2: In silico functional predictions of non-synonymous NPHS1 and NPHS2 variants detected in this study.
NPHS1-NPHS2 gene interaction
The Search Tool for Retrieval of Interacting Genes (STRING) database (version, 11.5) (https://string-db.org) was also used to predict functional interactions between proteins.
Statistical analysis
The paired t-test and one-way ANOVA with chi-square (χ2) values were applied to evaluate the demographic and clinical characteristics of NS cases using MedCalc (https://www.medcalc.org) and Social Science Statistics (https://www.socscistatistics.com/tests/chisquare2/default2.aspx) software.
Results
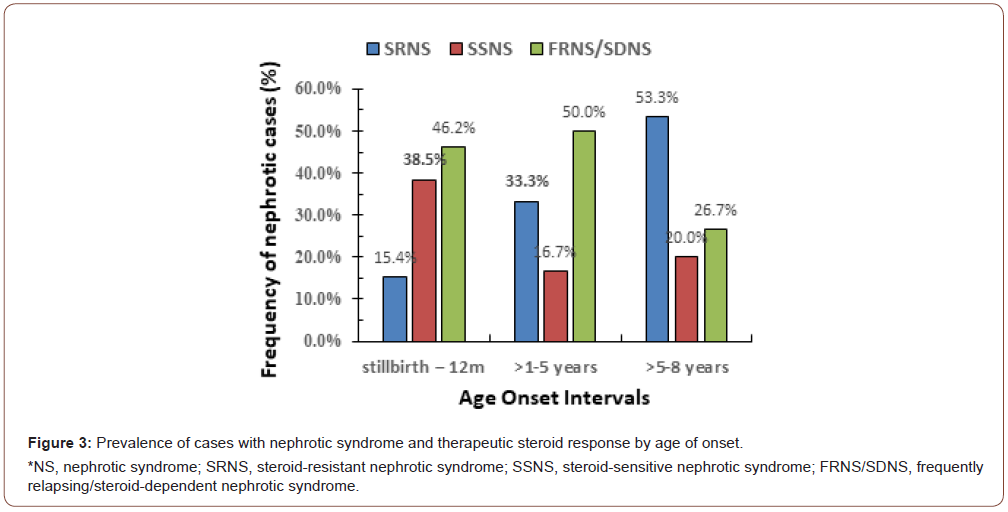
Among the 35 cases with NS, the female-to-male ratio was 13: 22 (i.e., 1:1.7), with no significant difference between genders (t-value = 0.329, 95% Cl, 4.1-6.3; P = 0.745). Additionally, no significant differences (P > 0.05) were found between cases and controls regarding age or gender. Among our cases, the mean age of onset was 3.0 ± 2.13 years (range, stillbirth-8 years). Six cases (17.1%) had an age of onset between stillbirth and 12 months, 21 cases (60%) had an age of onset >1-5 years, and 8 (22.9%) had an age of onset >5-8 years. Cases with FRNS/SDNS (46.2%) had a significantly younger age at presentation than cases with SSNS (38.5%) or SRNS (15.4%) (Figure 3).
Allelic variants of NPHS1 and NPHS2
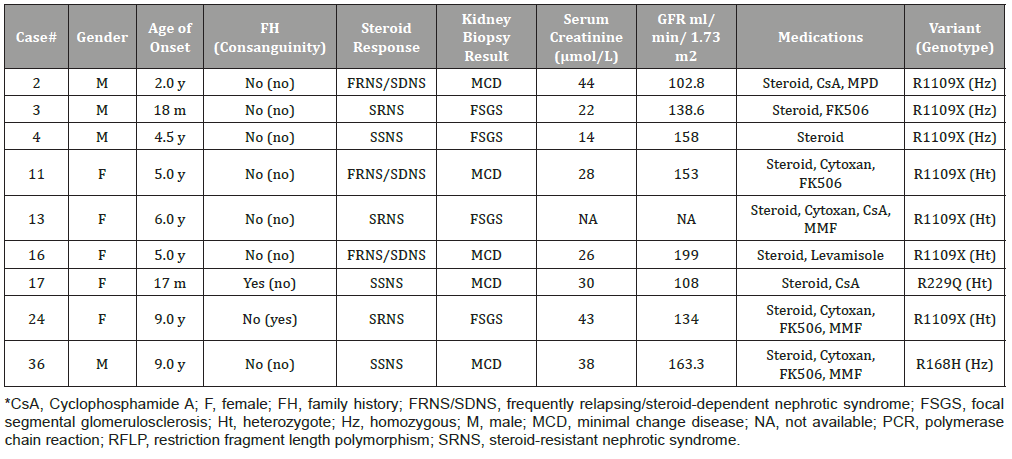
Our study found no cases due to the frameshift L41fsNX50 (c.121_122delCT) variant within the NPHS1 gene. Moreover, ten alleles leading to nonsense R1109X (c.3325C>T) variants were observed in exon 26 (Table 3). R1109X variants were associated with three homozygous NS cases (cases #2, #3, and #4) and four heterozygous NS cases (#11, #13, #16, and #24). Thus, the allele frequency of the R1109X variant in NPHS1 reached 14% (10/70 alleles). As for NPHS2, none of the cases with NS had the R291W (c.871C>T) variant. However, we detected the missense R168H (c.503G>A) variant in exon 4 and the R229Q (c.686G>A) variant in exon 5. The allele frequency of these NPHS2 variants was 4.3% (3/70 alleles).
Table 3: Clinical data of nephrotic cases with the NPHS1 and NPHS2 variants.
Response to steroid therapy
Among the NS cases, 12 (34.3%) were SRNS, 8 (22.9%) were SSNS, and 15 (42.9%) were FRNS/SDNS. (Figure 3) presents the age of onset of the cases and their responses to standard steroid therapy. Cases with FRNS/SDNS were significantly more likely to have an early age of onset stillbirth-12 months, or >1-5 years (46.2% or 50.0%, respectively) than were those with SSNS (38.5% or 16.7%) or SRNS (15.4% or 33.3%). In contrast, SRNS cases were more likely to have a later age of onset (>5-8 years) than were SSNS or FRNS/SDNS cases (53.3% vs. 20.0% vs. 26.7%, respectively). The response to steroid therapy in SRNS cases versus SSNS+FRNS/SDNS cases showed a χ2 = 4.1 (P = 0.044) for age of onset stillbirth-12 months, χ2 = 9.3 (P = 0.0023) for age of onset >1-5 years, and χ2 = 0.19 (P = 0.663) for >5-8 years.
Kidney biopsy outcome
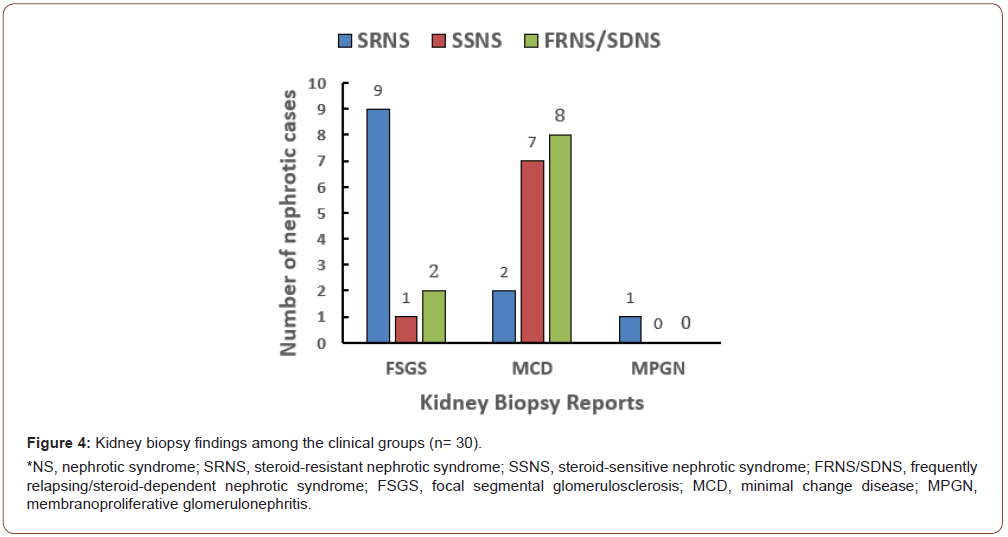
Kidney biopsy results were reported for 30 (85.7%) of 35 cases: 17 cases (56.7%) with MCD, 12 cases (40%) with FSGS, and 1 case (3.3%) with membranoproliferative glomerulonephritis (MPGN). MCD was most frequently reported in FRNS/SDNS cases (17/30 cases, 56.7%), whereas FSGS was the predominant histological phenotype in SRNS cases (9/12 cases, 75%) (Figure 4).
Genotype-phenotype correlation
(Table 3) represents the phenotype-genotype correlations between NS phenotypes and NPHS1/NPHS2 variants. Among cases with NPHS1 variants, three (cases #3, #13, and #24) had SRNS, three (cases #2, #11, and #16) had FRNS/SDNS, and one (case #4) had uncomplicated SSNS. Clinical and kidney biopsy analyses were used to diagnose four cases with FSGS and three cases with MCD.
Among the cases with NPHS2 variants, two cases were SSNS. Case #36, who was homozygous for the R168H variant, was a boy with no history of kidney disease. At nine years old, he was diagnosed with MCD using kidney biopsy analysis. His serum creatinine concentration was 38 μmol/L (normal range, 35-53 μmol/L), and his GFR was 163 ml/min/1.73 m2. There was no family history, and the parents were not consanguineous. His condition was treated with standard steroid medications: cyclophosphamide, FK506®, and MMF®, and described as a steroid-sensitive response (Table 3). Another patient carrying the NPHS2 R229Q variant (case #17) was a 17-month-old girl with a positive family history of proteinuria and kidney disease with no consanguinity of parents. A kidney biopsy diagnosed her as having MCD. Her condition was steroiddependent and treated with cyclosporine A. Her serum creatinine was 30 μmol/L at onset, and her GFR was 108 ml/min/1.73 m2.
NPHS1-NPHS2 Protein interactions
The protein-protein interaction (PPI) network of the NPHS1- NPHS2 protein-coding gene loci created by STRING software (https://string-db.org/) showed significantly more interactions than would be expected for a random set of proteins of the same size and degree of distribution drawn from the genome (PPI enrichment P-value = 0.00895) (Figure 5). More extended PPI network of the NPHS1-NPHS2 gene loci, including FYN, NCK1, TRPC6, CD2AP, and WASL protein-coding gene loci, were strong significant associations (PPI enrichment P-value = 2.64x10-5) (Table 4).
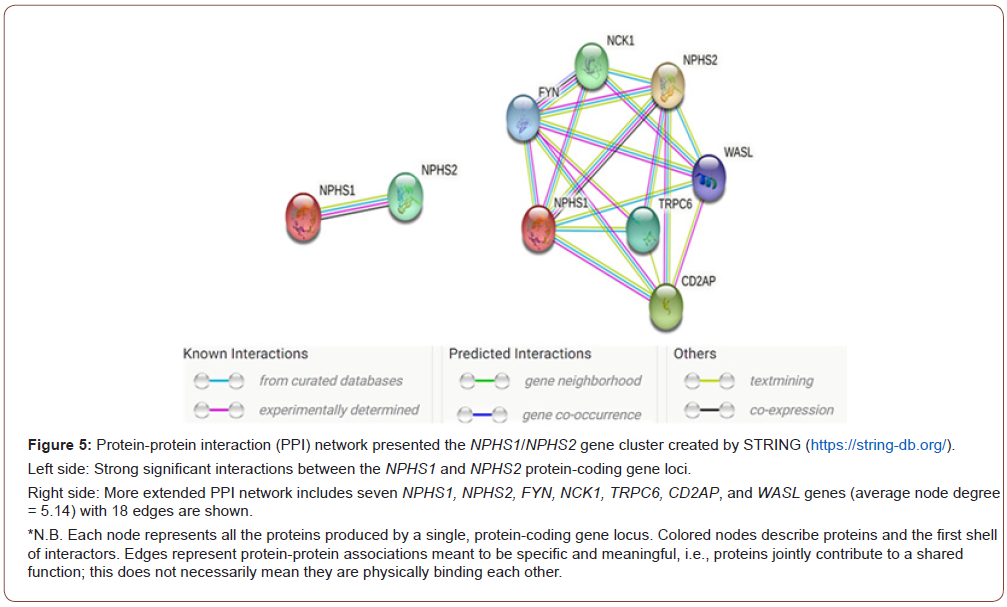
Table 4: Functional enrichment in NPHS1-NPHS2 protein-coding gene loci Network.

Discussion
In the present study, we explored variants in the NPHS1 (nephrin) and NPHS2 (podocin) genes and their association with disrupting the slit diaphragm architecture in the pathogenesis of earlier-onset NS. One known nonsense variant (R1109X) in NPHS1 and two missense variants (R168H and R229Q) in NPHS2 were reported here. Based on kidney biopsy reports, the identified variants were associated with MCD, FSGS, and MPGN phenotypes.
Based on electron microscopy analysis, abnormalities of the slit diaphragm have been seen in NS cases with MCD as the most common form of SSNS [36]. In addition, Horinouchi, et al. [37], have reported reduced podocin expression in patients with FSGS but not in those with MCD, suggesting that podocin expression and function in the slit diaphragm may not be altered in MCD.
Although NPHS1 variants were first reported in congenital NS of the Finnish population, subsequent studies have confirmed that NPHS1 is also a causative gene or susceptibility gene for kidney diseases such as FSGS, MCD with NS, IgA nephropathy, and MPGN [14,18-21,38]. The L41Nfs50 and R1109X variants in NPHS1 occur in the Finnish population at 78% and 16%, respectively[13], but rarely in non-Finnish ethnic populations[14]. Interestingly, 14% of NS cases carried the R1109X (Fin-minor) variant in our sample, but none carried the L41NfsX50 (Fin-major) variant. About 150 NPHS1 variants have been described among NS patients in different non- Finnish ethnic populations, including small insertions and deletions (indels), nonsense variants, and splicing variants [20,23,38-42]. Mostly, missense variants result in retention of nephrin in the endoplasmic reticulum, leading to complete loss of nephrin from the cell surface [43]. In addition, some reports have described silent variants that cause specific phenotypes by influencing mRNA structure or inactivating genes that affect gene splicing machinery and lead to exon skipping [44].
In Chinese cases with sporadic SRNS, synonymous changes in the NPHS2 gene (e.g., T741T, V763V, S1105S, A318A, and L346L) were observed in cases and healthy individuals, although there were no significant differences between cases and controls for some of these changes [44,45]. Large cohorts with familial SRNS in Europeans and North Americans have shown detection rates of 38% and 26% for homozygous and compound heterozygous NPHS2 variants, respectively, but 6-19% for NPHS2 variants in sporadic NS cases [8,28]. Two studies have confirmed that cases with FSGS and two pathogenic NPHS2 variants are generally characterized by early-onset disease, response to SRNS treatment, and reduced risk of FSGS recurrence after kidney transplantation [28]. The R229Q and R291W variants in NPHS2 were previously reported as compound heterozygotes [46,47], detected in 22 (84.6%) of 26 SRNS cases, 42.3% of whom went on to develop end-stage kidney disease (ESKD) [28].
Although only one compound heterozygous R229Q variant (1/70; 1.4%) was detected in the present study, Mikó, et al. [48]. Described this variant as the most frequent NPHS2 variant in the general population and as the first human variant for which pathogenicity depends on another associated allele. The R229Q variant can cause FSGS if it is trans-associated with specific variants that affect the protein region spanning 270-351 residues [48]. The coexistence of the R229Q variant and other variants in residues 260 and 310 has been reported in three children who developed ESKD [49]. More recently, Righetti, et al. [50]. concluded that compound heterozygous R229Q or R291W variants might be associated with the FSGS phenotype, but neither heterozygous variant was associated with significant proteinuria.
Conclusion
Our study demonstrated that the nonsense R1109X variant of the Finnish type in the NPHS1 gene is present in NS patients with sporadic FSGS and MCD in the Saudi community. NPHS1 variants could represent an important role in the pathogenesis of these diseases, but more research is needed to explain the physiopathological mechanisms of these variants. Moreover, nearly 50% of Saudi cases at younger ages with FRNS/SDNS were still challenging during repeated protocols of corticosteroids and second-line immunosuppressive agents associated with morbidity. Although the allele frequency of the NPHS2 gene in cases with NS was 4.3% (3 of 70 cases) in our study, the sample size was relatively small. During the preparation of this manuscript, external exome sequencing for 73 of our NS cases revealed two additional previously unreported variants that are yet to be confirmed in NPHS1 and NPHS2. Thus, extending our research studies using exome sequencing could help expedite the discovery of NS genetic heterogeneity among the Saudi community.
Acknowledgment
The authors would like to thank the patients for sharing in this study. We are also grateful to Drs. Burhan Edrees (Department of Pediatrics, UQU) and Khawla Rahim (King Fahd Medical Center) allowed us to study their patients.
Conflict of Interest
The authors report no conflicts of interest in this work.
References
- Eddy AA, Symons JM (2003) Nephrotic syndrome in childhood. Lancet (23) 362: 629-639.
- Tryggvason K, Patrakka J, Wartiovaara J(2006) Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med (354): 1387-401.
- Holmberg C, Antikainen M, Ronnholm K, Ala Houhala M, Jalanko H(1995) Management of congenital nephrotic syndrome of the Finnish type. Pediatr Nephrol 9(1): 87-93.
- Seppala M, Rapola J, Huttunen NP, Aula P, Karjalainen O, Ruoslahti E(1976) Congenital nephrotic syndrome: prenatal diagnosis and genetic counselling by estimation of aminotic-fluid and maternal serum alpha-fetoprotein. Lancet 2: 123-125.
- Kim JS, Bellew CA, Silverstein DM, Aviles DH, Boineau FG, et al. (2005) High incidence of initial and late steroid resistance in childhood nephrotic syndrome. Kidney Int 68 (3): 1275-1281.
- Kari JA, Alhasan KA, Albanna AS, Safdar OY, Shalaby MA, et al. (2020) Rituximab versus cyclophosphamide as first steroid-sparing agent in childhood frequently relapsing and steroid-dependent nephrotic syndrome. Pediatr Nephrol 35(8): 1445-1453.
- Larkins NG, Liu ID, Willis NS, Craig JC, Hodson EM (2020) Non-corticosteroid immunosuppressive medications for steroid-sensitive nephrotic syndrome in children. Cochrane Database Syst Rev 16(4): CD002290.
- Fuchshuber A, Gribouval O, Ronner V, Kroiss S, Karle S, et al. (2001) Clinical and genetic evaluation of familial steroid-responsive nephrotic syndrome in childhood. J Am Soc Nephrol 12(2): 374-378.
- D Agati VD, Kaskel FJ, Falk RJ (2011) Focal segmental glomerulosclerosis. N Engl J Med. 365(25): (2398-411).
- De Vriese AS, Sethi S, Nath KA, Glassock RJ, Fervenza FC (2018) Differentiating Primary, Genetic, and Secondary FSGS in Adults: A Clinicopathologic Approach. J Am Soc Nephrol 29(3): 759-774.
- Vijayan P, Hack S, Yao T, Qureshi MA, Paterson AD, et al. (2021) LAMA2 and LOXL4 are candidate FSGS genes. BMC Nephrol 22(1): 320.
- Buscher AK, Beck BB, Melk A, Hoefele J, Kranz B, et al. (2016) Rapid response to cyclosporin A and favorable renal outcome in nongenetic versus genetic steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 11(2): 245-253.
- Kestila M, Lenkkeri U, Mannikko M, Lamerdin J, McCready P, et al. (1998) Positionally cloned gene for a novel glomerular protein-nephrin-is mutated in congenital nephrotic syndrome. Mol Cell 1(4):575-582.
- Lenkkeri U, Mannikko M, McCready P, Lamerdin J, Gribouval O, et al. (1999) Structure of the gene for congenital nephrotic syndrome of the Finnish type (NPHS1) and characterization of mutations. Am J Hum Genet 64(1): 51-61.
- Martin CE, Jones N Nephrin (2018) Signaling in the Podocyte: An Updated View of Signal Regulation at the Slit Diaphragm and Beyond. Front Endocrinol (Lausanne) 5(9): 302.
- Verma R, Venkatareddy M, Kalinowski A, Li T, Kukla J, et al. (2018) Nephrin is necessary for podocyte recovery following injury in an adult mature glomerulus. PLoS One 13(6): e0198013.
- Huttunen NP (1976) Congenital nephrotic syndrome of Finnish type. Study of 75 patients. Arch Dis Child 51(5): 344-348.
- Hinkes BG, Mucha B, Vlangos CN, Gbadegesin R, Liu J, et al. (2007) Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics 119(4): (e907-19).
- Schoeb DS, Chernin G, Heeringa SF, Matejas V, Held S, et al. (2010) Nineteen novel NPHS1 mutations in a worldwide cohort of patients with congenital nephrotic syndrome (CNS). Nephrol Dial Transplant 25(9): 2970-2976.
- Heeringa SF, Vlangos CN, Chernin G, Hinkes B, Gbadegesin R, et al. (2008) Thirteen novel NPHS1 mutations in a large cohort of children with congenital nephrotic syndrome. Nephrol Dial Transplant 23(11): 3527-3533.
- Nguyen TK, Pham VD, Nguyen TH, Pham TK, Nguyen TQ (2017) Three novel mutations in the NPHS1 gene in Vietnamese patients with congenital nephrotic syndrome. Case Rep Genet 2357282.
- Fuchshuber A, Jean G, Gribouval O, Gubler MC, Broyer M, et al. (1995) Mapping a gene (SRN1) to chromosome 1q25-q31 in idiopathic nephrotic syndrome confirms a distinct entity of autosomal recessive nephrosis. Hum Mol Genet 4(11): 2155-2158.
- Beltcheva O, Martin P, Lenkkeri U, Tryggvason K. (2001)Mutation spectrum in the nephrin gene (NPHS1) in congenital nephrotic syndrome. Hum Mutat 17(5): 368-373.
- Mohey H, Thibaudin L, Laurent B, Berthoux F (2013) The podocin mutation R229Q and early recurrence (within the first year) of glomerular disease after renal transplantation. Ann Transplant 28(18): 436-442.
- Pereira AC, Pereira AB, Mota GF, Cunha RS, Herkenhoff FL, et al. (2004) NPHS2 R229Q functional variant is associated with microalbuminuria in the general population. Kidney Int 65 (3) 1026-1030.
- Guaragna MS, Lutaif A, Maciel-Guerra AT, Belangero VMS, Guerra-Junior G,et al. (2017)NPHS2 mutations: A closer look to Latin American countries. Biomed Res Int 7518789.
- Winn MP. (2002) Not all in the family: mutations of podocin in sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol 13(2): (577-579.
- Caridi G, Bertelli R, Di Duca M, Dagnino M, Emma F, et al. (2003) Broadening the spectrum of diseases related to podocin mutations. J Am Soc Nephrol 14(5): 1278-1286.
- Tsukaguchi H, Sudhakar A, Le TC, Nguyen T, Yao J, et al. (2002) NPHS2 mutations in late-onset focal segmental glomerulosclerosis: R229Q is a common disease-associated allele. J Clin Invest 110: 1659-1666.
- Machuca E, Hummel A, Nevo F, Dantal J, Martinez F,et al. (2009) Clinical and epidemiological assessment of steroid-resistant nephrotic syndrome associated with the NPHS2 R229Q variant. Kidney Int 75(7): 727-735.
- Lu L, Wan H, Yin Y, Feng WJ, Wang M, et al. The p.R229Q variant of the NPHS2 (podocin) gene in focal segmental glomerulosclerosis and steroid-resistant nephrotic syndrome: a meta-analysis. Int Urol Nephrol 46(7): 1383-1393.
- Londeree J, McCracken CE, Greenbaum LA, Anderson EJ, Plantinga LC, et al. 2022) Estimation of childhood nephrotic syndrome incidence: data from the Atlanta metropolitan statistical area and meta-analysis of worldwide cases. J Nephrol 35(2): 575-583.
- Koskela M, Ylinen E, Ukonmaanaho EM, Autio-Harmainen H, Heikkila P, et al. (2017) The ISKDC classification and a new semiquantitative classification for predicting outcomes of Henoch-Schonlein purpura nephritis. Pediatr Nephrol 32(7): 1201-1209.
- Elhawary NA, Jiffri EH, Jambi S, Mufti AH, Dannoun A, et al. (2018) Molecular characterization of exonic rearrangements and frame shifts in the dystrophin gene in Duchenne muscular dystrophy patients in a Saudi community. Hum Genomics 12(1): 18.
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, et al. (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24(4): 349-354.
- Patrakka J, Lahdenkari AT, Koskimies O, Holmberg C, Wartiovaara J, et al. (2002) The number of podocyte slit diaphragms is decreased in minimal change nephrotic syndrome. Pediatr Res 52(3): 349-355.
- Horinouchi I, Nakazato H, Kawano T, Iyama K, Furuse A, et al. (2003) In situ evaluation of podocin in normal and glomerular diseases. Kidney Int 64(6): 2092-2099.
- Ismaili K, Pawtowski A, Boyer O, Wissing KM, Janssen F, et al. (2009) Genetic forms of nephrotic syndrome: a single-center experience in Brussels. Pediatr Nephrol 24: 287-294.
- Aya K, Tanaka H, Seino Y (2000) Novel mutation in the nephrin gene of a Japanese patient with congenital nephrotic syndrome of the Finnish type. Kidney Int 57(2): 401-404.
- Gigante M, Monno F, Roberto R, Laforgia N, Assael MB, et al. (2002) Congenital nephrotic syndrome of the Finnish type in Italy: a molecular approach. J Nephrol 15(6): 696-702. PMID: 12495287.
- Sako M, Nakanishi K, Obana M, Yata N, Hoshii S, et al. (2005) Analysis of NPHS1, NPHS2, ACTN4, and WT1 in Japanese patients with congenital nephrotic syndrome. Kidney Int (67): 1248-1255.
- Frishberg Y, Ben-Neriah Z, Suvanto M, Rinat C, Mannikko M, et al. (2007) Misleading findings of homozygosity mapping resulting from three novel mutations in NPHS1 encoding nephrin in a highly inbred community. Genet Med 9(3): 180-184.
- Liu XL, Done SC, Yan K, Kilpelainen P, Pikkarainen T, et al. (2004) Defective trafficking of nephrin missense mutants rescued by a chemical chaperone. J Am Soc Nephrol 15(7): 1731-1738.
- Yu Z, Ding J, Huang J, Yao Y, Xiao H, et al. (2005) Mutations in NPHS2 in sporadic steroid-resistant nephrotic syndrome in Chinese children. Nephrol Dial Transplant 20(5): 902-908.
- Mao J, Zhang Y, Du L, Dai Y, Gu W, et al. (2007) NPHS1 and NPHS2 gene mutations in Chinese children with sporadic nephrotic syndrome. Pediatr Res 61(1): 117-122.
- Tory K, Menyhard DK, Woerner S, Nevo F, Gribouval O, et al. (2014) Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 46: 299-304.
- Straner P, Balogh E, Schay G, Arrondel C, Miko A, et al. (2018) C-terminal oligomerization of podocin mediates interallelic interactions. Biochim Biophys Acta Mol Basis Dis 1864(7): 2448-2457.
- Miko A, D KM, Kaposi A, Antignac C, Tory K (2018) The mutation-dependent pathogenicity of NPHS2 p.R229Q: A guide for clinical assessment. Hum Mutat 39(12): 1854-1860.
- Feltran LS, Varela P, Silva ED, Veronez CL, Franco MC, et al. (2017) Targeted next-generation sequencing in Brazilian children with nephrotic syndrome submitted to renal transplant. Transplantation 101(12): 2905-2912.
- Riguetti MTP, Varela P, Fernandes DE, Polito MG, Casimiro FM, et al. (2020) Familial focal segmental glomerulosclerosis with late-onset presentation and R229Q/R291W podocin mutations. Front Genet 11: 533373.
-
Ahmad O Babalghith, Nasser A Elhawary, Iman S Abumansour, Ehab M Melibary Ghydda Alghamdi, Ikhlas A Sindi, et al., NPHS1 and NPHS2 Variants Associated with Early-Onset Frequently Relapsing or Steroid-Dependent Nephrotic Syndrome in the Saudi community. Annal Urol & Nephrol. 3(2): 2022. AUN.MS.ID.000557.
-
Dialysis, Education, Knowledge, Nephrologists, Transplantation, Chronic kidney disease
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.