Research Article
Research Article
A Novel Gene and Comparison of Gene Expressions in Low- and High-Grade Renal Cell Carcinomas
Yasemin Yuyucu Karabulut1*, Funda Bozkurt1, Oznur Bucak1, Emre Cagatay Kose1, Bahar Taşdelen 2, Ali Nebioglu3, Hasan Erdal Doruk 3, Mehmet Emin Erdal 4
1Department of Pathology, Mersin University, Turkey
2Department of Biostatistics, Mersin University, Turkey
3Department of Urology, Mersin University, Turkey
4 Department of Medical Biology and Genetics, Mersin University, Turkey
Yasemin Yuyucu Karabulut, Department of Pathology, Mersin University, Turkey
Received Date: April 04, 2023; Published Date: April 27, 2023
Abstract
YYK: 0000-0001-6619-6868
FB: 0000-0003-1407-5655
OB: 0000-0002-5778-9079
EcK: 0000-0002-6903-9064
BT: 0000-0001-8146-4912
AN: 0000-0001-6325-1534
HED: 0000-0001-5671-9602
MEE: 0000-0002-6191-2930
Background/Aim: Determining the genetic characteristics of renal cell carcinoma (RCC) and their genetic differences will make a significant contribution and guide in the diagnosis of the disease, determination of prognosis and survival, and targeted therapies. We aim to determine and compare the expressions of DKK1, sFRP2, MALAT1, PMCA1, and TRPV6 genes in renal cell carcinomas and correlate them with patient survival.
Methods: As a prospective cohort study, 150 patients diagnosed with RCC in nephrectomy specimens and 40 non-tumoral renal parenchymal tissues evaluated between 2008 and 2018 in our center were included in the study. Tumor type, nucleolar grade and pathological stage interactions were evaluated using factorial ANOVA and a post-hoc planned contrast test. The effect of gene expression on patient survival was evaluated with the Cox Proportional Hazards model.
Results: The difference between the patient and healthy groups in terms of DKK1, SFRP2, MALAT1 and PMCA1 gene expressions was found to be statistically significant. DKK1 and SFRP2 gene expression were higher in healthy tissues (P < 0.001, P < 0.001). MALAT1 and PMCA1 gene expression were found to be higher in tumor tissues (P = 0.011, P = 0.046). The expressions of SFRP2, MALAT1 and TRPV6 in tumors with low WHO/ ISUP nucleolar grade were found to be significantly higher (P < 0.001, P = 0.001, P < 0.001). PMCA1 expression was found to be higher in RCCs with nucleolar grades 3 and 4 (P = 0.016). PMCA1 expression was also found to be associated with longer survival and lower death rates (P = 0.007).
Conclusions: The higher expression and relatively protective effect of PMCA1 in RCC may lead to the determination of new molecular targets in the diagnosis and treatment of RCC.
Keywords: Gene expression, Renal cell carcinoma, PMCA1, Nephropathology, Kidney
Background
Renal cell carcinoma (RCC) is a common malignant tumor that constitutes approximately 85% of all kidney malignancies [1]. If RCCs are detected while limited to the kidney capsule, curative treatment is usually possible [2]. Although the number of RCC cases detected incidentally by imaging methods has increased today, metastatic lesions can be detected in approximately 30% of patients at the time of diagnosis [3]. For this reason, a better understanding of the molecular and genetic characteristics of RCC will make a significant contribution to the detection of the disease, the determination of the prognosis, and selecting the targeted treatment regimen. The Wnt/beta-catenin signaling pathway affects the development mechanism of RCCs. They participate in a variety of cellular processes, including tissue homeostasis maintenance, cell growth control, embryogenesis, and oncogenesis [4]. Members of the Wnt antagonist family include secreted Frizzled-related protein (sFRP), Wnt inhibitory factor 1 (Wif1), Xenopus Cerberus, Wise, and Dickkopf (DKK) proteins. Functional loss of Wnt antagonists contributes to the activation of the Wnt pathway. This situation may cause carcinogenesis by dysregulation of cell proliferation and differentiation [5].
Studies have found that DKK1 expression in RCC cells is significantly reduced [6]. Urakami et al. showed that methylation levels of Wnt antagonists including sFRP1, sFRP2, sFRP4, sFRP5, WIF1, and DKK3 were higher in RCC tissues than in normal renal tissues [5]. Since the expression of Wnt antagonists in many cancer types is suppressed by promoter hypermethylation, it has been suggested that these proteins have tumor suppressor properties [7]. It is known that long non-coding RNAs (lncRNA) play a role in tumor development. LncRNAs are RNA molecules longer than 200 nucleotides with a limited protein-coding ability [8]. They function in various biological processes such as proliferation, apoptosis, cell migration, and invasion [9]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is an 8000 nucleotide lncRNA that is commonly found and expressed in many locations. Studies have found that the expression of MALAT1 in various tumors is increased and this increase is associated with metastasis and recurrence [8]. As a result of the study by Zhang et al., MALAT1 expression was found to be higher in RCC tissues than in non-tumor tissues, and it was found that survival time was shorter among RCC patients with higher expression levels [10]. Plasma membrane Ca2+ ATPase (PMCA) is the main regulator of free intracellular calcium and plays a role in maintaining homeostasis. Today, four isoforms of PMCA, 1, 2, 3, and 4, have been defined [11, 12]. PMCA1 and 4 are found in most cell types, while PMCA2 and 3 are distributed in a limited number of tissues [13]. In a study, it was found that PMCA1 expression decreased in oral squamous cell carcinomas [11]. However, regarding RCCs, it is not known whether PMCA1 plays a role in carcinogenesis. TRP channels form a large and functionally versatile superfamily of cation channel proteins expressed in many cell types [14]. TRPV6, a member of the TRP family, is a channel with high Ca2+selectivity. At the same time, cellular Ca2+channels are effective in the induction and regulation of apoptosis and are associated with the development of cancer [15, 16].
Our aim in this study is to determine the expression of DKK1, SFRP2, MALAT1, PMCA1 and TRPV6 in low and high nuclear grade RCCs and to evaluate the expression difference between these groups. It is thought that determining the genetic characteristics of RCCs and their genetic differences will make a significant contribution and guide in the diagnosis of the disease, determination of prognosis and targeted therapies.
Materials and methods
Selection of Cases
150 patients diagnosed with RCC in nephrectomy specimens evaluated between 2008 and 2018 in Mersin University Faculty of Medicine, Department of Medical Pathology, and as control 40 non-tumoral renal parenchymal tissues that were resected for non-tumoral reasons, were included in the study. Histological diagnosis was determined according to the WHO guideline. WHO/ISUP nucleolar grade 1 and 2 tumors are grouped as low grade, 3 and 4 as high grade. The cases were determined as 75 cases with low grades and 75 cases with high grades. Pathological tumor stages (pT) 1 and 2 were categorized as low stages; those with (pT) 3 and 4 were classified as high stages.
Molecular Genetic Analysis
RNA Isolation: 10 micron-thick sections were taken from paraffin block tissue materials of 150 patients diagnosed with RCC and 40 healthy cases. After the deparaffinization process, total RNA isolation was achieved by the trizol method.
Complementary DNA (cDNA) Synthesis: 15μl reverse transcriptase reaction was prepared as a mix of; 5μl total RNA, 0,5 μg Oligo (dT) 15 Primer, 1 × RT buffer, dNTP 0,25mM, 50 units modified M-MuLV Reverse Transcriptase (Thermo Scientific), 25 units RiboLock RNase inhibitor (Thermo Scientific) and nuclease-free water. cDNAs were obtained in the reaction mixture by performing Reverse transcriptase PCR (RT-PCR) using Thermal Cycler (Techne Flexigene, Cambridge, UK) conditions set as 1 cycle at 16 ° C for 30 minutes, 42 ° C for 30 minutes, 85 ° C for 5 minutes.
Quantitative-comparative CT (ΔΔCT) Real-time PCR
Expression analyzes were performed using the ABI Prism 7500 (quantitative-comparative CT) Real-Time PCR System (Applied Biosystems) with SDS 2.0.6 software program. Specific primers and fluorogenic probes were designed for mRNAs with the Primer Express 3.0 software (Applied Biosystems) (Metabion-International AG, D-82152 Martinsried/Deutschland). Homo sapiens actin, beta (ACTB) was used as the endogenous control gene and the RNA mixture obtained from RNA from the control group was used as the reference RNA sample. 25μl expression mix was prepared to contain; 3 μl of RT-PCR product 12,5 μl of 2X TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nmol of each primer (Primer F and Primer R) and 200 nmol TaqMan® probes. The analysis was performed with the execution method prepared as 2 minutes 1 cycle at 50 ° C, 1 cycle for 10 minutes at 95 ° C, 15 seconds at 95 ° C and 50 cycles for 1,5 minutes at 60 ° C. The results were quantitatively determined by the ΔΔCT method in the Real-Time PCR device. Data normalization was achieved by calculating the 2-ΔΔCTvalue.
Statistical analysis
Statistical analyzes were made with the STATISTICA Version 13.3 program. Data are summarized as mean, standard deviation, number and percentage. Gender distributions of the patient and healthy groups were compared with the Chi-square test and age distributions with the independent t-test. Normal distribution control of gene expression levels was done with the Shapiro Wilk’s test. Tumor type, nucleolar grade, pathological stage interactions were evaluated using factorial ANOVA and a post-hoc planned contrast test. The effect of gene expression on patient survival was evaluated with the Cox Proportional Hazards model. The situations where the P-value was <0.05 were considered statistically significant.
Results
This study includes 150 cases with an RCC diagnosis. As a control group, 40 healthy, tumor-free cases were included in the study. The clinicopathological characteristics of the cases are summarized in (Table 1).The difference between the patient and healthy groups in terms of DKK1, SFRP2, MALAT1 and PMCA1 gene expression was found to be statistically significant. There was no significant difference between non-tumoral tissues and tissues with RCC in terms of TRPV6 gene expression (P = 0.066). It was found that DKK1 and SFRP2 gene expression was higher in healthy tissues and their expression decreased in tumor tissues (P < 0.001, P < 0.001). MALAT1 and PMCA1 gene expression were found to be higher in tumor tissues than in healthy tissues (P = 0.011, P = 0.046) (Table 2). In terms of all genes, no significant expression difference was found between clear cell RCC and papillary type RCC (P > 0.05). Relationship between DKK1, SFRP2, MALAT1, PMCA1, TRPV6 gene expressions and clinicopathological factors in renal cell carcinomas. The expressions of SFRP2, MALAT1, and TRPV6 in tumors with low WHO/ISUP nucleolar grade were found to be significantly higher than in tumors with a high nucleolar grade (P < 0.001, P = 0.001, P < 0.001). PMCA1 expression was found to be higher in RCCs with nucleolar grades 3 and 4 (P = 0.016) (Table 3). PMCA1 expression was also found to be associated with longer survival and lower death rates (P = 0.007).
Table 1:Clinicopathological features of the patients with renal cell carcinoma and the controls.
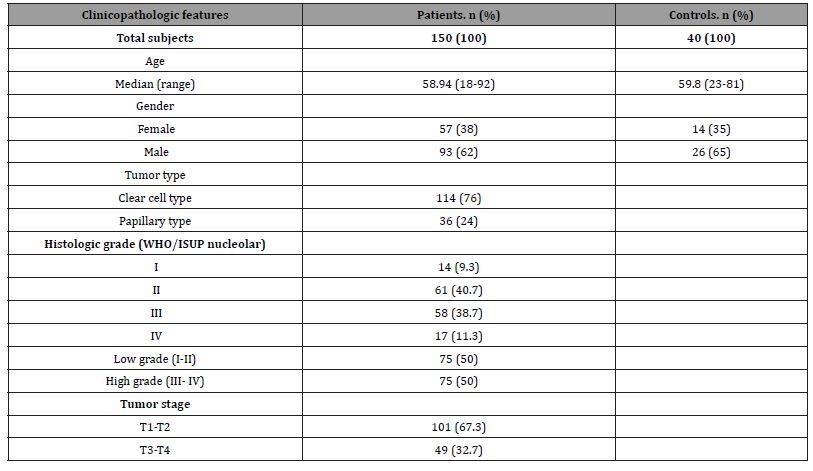
Table 2:Comparison of gene expressions in tumor tissues and normal tissues
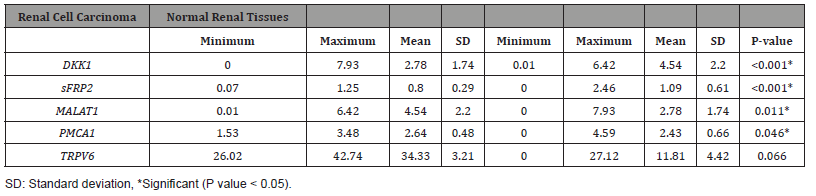
Table 3:Comparison of gene expressions in low- and high-grade renal cell carcinomas
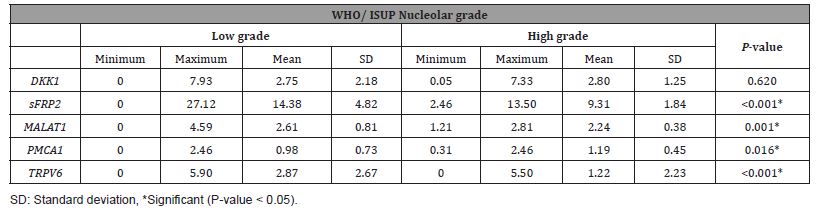
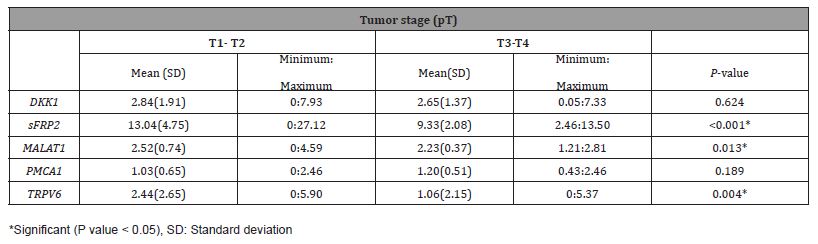
Table 4:Comparison of gene expressions in low and high stage renal cell carcinomas.
It was found that SFRP2, MALAT1 and TRPV6 gene expressions were higher in clear cell RCC compared to tumors with low WHO/ ISUP nucleolar grade (P < 0.001, P = 0.006, P = 0.012). The expression of PMCA1 was found to be lower in low nucleolar grade tumors than in high nucleolar grade tumors (P = 0.024). In papillary type RCC, SFRP2 and TRPV6 genes were found to be significantly more expressed in low WHO/ISUP nucleolar grade tumors than in high nucleolar grade (P = 0.001, P = 0.001).
Significant differences were found between clear cell and papillary type RCCs with a high nucleolar grade in terms of DKK1 (P = 0.049) and the TRPV6 gene (P = 0.024). Besides, it was observed that DKK1 and TRPV6 genes were more expressed in high WHO/ ISUP nucleolar grade clear cell RCC than in papillary type RCC. It was determined that MALAT1, SFRP2 and TRPV6 gene expressions were higher in tumors with low pT, and their expression decreased in pT stage 3-4 tumors (P = 0.013, P < 0.001, P = 0.004) (Table 4).
SFRP2 and MALAT1 gene expressions were found to be significantly higher in the low pT group in clear cell RCC (P = 0.002, P = 0.032). In papillary type RCC, a significant difference was found between tumors with low and high pT in terms of SFRP2 (P < 0.001) and TRPV6 (P < 0.001) gene expressions. Gene expressions in tissues with low pathological tumor stage clear cell and papillary type RCC were compared, and a statistically significant difference was found in terms of the SFRP2 gene (P = 0.036). SFRP2 gene expression was higher in the papillary type than in the clear cell type in RCCs with low pT. A statistically significant difference was found between the high pathological tumor stage clear cell and papillary type RCC in terms of the TRPV6 gene (P = 0.016). TRPV6 gene expression is significantly higher in the clear cell type than in the papillary type in high-stage tumors.
Discussion
Despite advances in the diagnosis and treatment of RCCs, the prognosis is poor, metastasis is observed in approximately onethird of the patients at the time of diagnosis [17]. Detection of molecular mechanisms and differences that are effective in the pathogenesis and progression of RCCs can make a significant contribution to the early diagnosis and treatment of the disease. In the study of Hiroshi et al., it was found that DKK1 expression was significantly suppressed in clear cell type RCC cells and it was shown that DKK1 re-expression induced apoptosis in renal cancers [6]. In studies on gastrointestinal cancers, it has been found that the DKK family suppresses the growth of cancer cells, and their level in cancer cells has been shown to decrease [18]. Inhibition of the SFRP2 gene by hypermethylation has been reported in stomach, breast, renal and colon cancers [19]. In various studies, it was found that the methylation of the DKK1 gene and other Wnt antagonists was higher in high pT and grade tumors, and it was found that the DKK1 level was decreased in high-grade RCCs [5, 6]. Similarly, in our study, it was observed that DKK1 expression was higher in healthy tissues and its expression in RCC tissues was found to be significantly reduced. When the clear cell and papillary type RCC tissues with high nucleolar levels were compared, it was found that the expression of DKK1 in the clear cell type was higher than in the papillary type.
The expression of the SFRP2 gene, another Wnt antagonist, was found at a lower level in the tumors in our study. It was found that SFRP2 gene expression was significantly decreased in tumors with high pT and nucleolar grade compared to low-grade tumors. The data obtained from our study and previous studies support the view that DKK1 and SFRP2 have tumor suppressor properties and have a significant effect on the pathogenesis and progression of RCCs.
It is known that LncRNAs have an important role in cancer development and progression. MALAT1 is a nuclear IncRNA expressed on chromosome 11q13, also called nuclear-enriched transcript 2 (NEAT2). Previous research has shown that MALAT1 is highly expressed in various cancers such as lung, breast, liver, pancreatic, colon, and oral squamous cell cancer [20-23]. In the first study on clear cell RCCs, it was found that the lncRNA MALAT1 level was higher in the tumor than in non-tumor tissues. A correlation was found between MALAT1 expression level and tumor size, tumor stage, and lymph node metastasis, but not with histological grade and distant metastasis [24]. In the study of Li et al., MALAT1 levels were found to be high in RCC tissues, and it was found that suppression of the gene significantly inhibited the proliferation and migration of RCC cells [25]. In our study, it was determined that MALAT1 expression increased significantly in tumor cells. We also noticed the expression of MALAT1 in the clear cell type was high in tumors with low pT and histological grade, but no significant relationship was found between these parameters in the papillary type. Although MALAT1 expression is high in RCCs, its role in tumor proliferation and metastasis is not known clearly. Somehow, MALAT1’s significant increase in RCC tissues and suppression of its expression decreases cell proliferation, which may make it a promising candidate in therapy.
calcium homeostasis, transporting calcium across the plasma membrane by active transport. There are four different isoforms (PMCA1-4) encoded by four genes. While PMCA1 and PMCA4 are expressed in many cells, PMCA2 and PMCA3 have a limited tissue distribution [11-13]. In recent studies, an increase in PMCA1 expression was found in breast cancer, rhabdomyosarcoma, and hepatocarcinoma [26-28]. In a study evaluating PMCA expression in colon cancer, a relationship was found between differentiation in cancer cells and the level of PMCA4. However, it has been found that there is no significant change in PMCA1 expression in colon cancer [28]. In the study of Saito et al., it was observed that PMCA1 expression was lower in oral squamous cell carcinomas. PMCA1 gene inactivation is thought to be a frequent and early event during oral carcinogenesis. However, no relationship was found between PMCA1 expression and tumor stage or grade [11]. Regarding RCCs, it is not yet known whether the PMCA1 gene is associated with carcinogenesis. Our study is the first and unique study evaluating PMCA1 expression in RCC cells, and it was found that PMCA1 expression was higher in RCC cells than in normal healthy renal tissue cells. Besides, it was noted that the expression showed a marked increase in tumors of WHO/ISUP nucleolar grade (3-4). PMCA1 expression was also found to be associated with longer survival and lower death rates. In summary, it was concluded that PMCA1 expression is increased in RCC tissues, increased with higher grades, and associated with better survival. Cellular Ca2+channels are important in the regulation of apoptotic pathways and are associated with cancer development. TRPV6 is a highly selective channel responsible for maintaining intracellular Ca2+levels. TRPV6 levels have been investigated in various types of cancer, but promising results have been obtained, especially in prostate and colon carcinomas [29]. In the study of Wu et al., TRPV5/TRPV6 was found to be expressed in normal human kidney and RCC tissues, but its expression was found to be decreased in RCC [30]. In our study, TRPV6 expression was detected in normal renal tissue and RCC tissues, but although there was no significant difference between them, it was found that the expression was high in tumors with low WHO/ISUP nucleolar grade and pT, and expression decreased as the tumor progressed. The effect of TRPV6 on RCC carcinogenesis is still uncertain.
Conclusion
In our study, we examined the expressions of DKK1, SFRP2, MALAT1, PMCA1, and TRPV6 genes both in normal renal tissue and in RCCs. Although our results are largely compatible with the current literature, the main point that makes our study significant and different is that it is the first and unique study evaluating PMCA1 expression in RCC cells. PMCA1 expression was found to be higher in RCC cells than in normal healthy renal parenchyma cells, most prominently in WHO/ISUP nucleolar grade 3-4 RCCs. The results obtained, especially for PMCA1, will lead to the determination of new molecular targets in the diagnosis and treatment of RCC.
Limitations
In the current literature there is not a similar paper so comparing all of our results properly could not be possible. Further studies, with larger case series, are needed to explain the effects of these genes in RCC.
Acknowledgements
None
Conflict of Interest
Author declares no conflict of interest
References
- Jonasch E, Gao J, Rathmell WK (2014) Renal cell carcinoma. BMJ 10; 349: g4797.
- Lerner SE, Hawkins CA, Blute ML, Grabner A, Wollan PC, et al., (1996) Disease outcome in patients with low stage renal cell carcinoma treated with nephron sparing or radical surgery. J Urol 155(6):1868-73.
- Bukowski RM (1997) Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin-2 Cancer 1; 80(7):1198-220.
- Cohen ED, Tian Y, Morrisey EE (2008) Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development135(5): 789-798.
- Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K (2006) Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res 1;12(23): 6989-6997.
- Hirata H, Hinoda Y, Nakajima K, Kawamoto K, Kikuno N et al. (2011)Wnt antagonist DKK1 acts as a tumor suppressor gene that induces apoptosis and inhibits proliferation in human renal cell carcinoma. Int J Cancer 15;128(8):1793-1803.
- Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, et al. (1997) A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci U S A 1; 94(7): 2859-2863.
- Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014 12; 25(5): 666-681.
- Yu G, Yao W, Gumireddy K, Li A, Wang Jet al. (2014)Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther 13(12): 3086-3097.
- Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH (2015) Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol 36(4): 2947-2955.
- Boczek T, Lisek M, Ferenc B, Kowalski A, Stepinski D, Wiktors et al. (2014) Plasma membrane Ca2+-ATPase isoforms composition regulates cellular pH homeostasis in differentiating PC12 cells in a manner dependent on cytosolic Ca2+ elevations. PLoS One 11; 9(7): e102352.
- Strehler EE, Zacharias (2001)DA Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev 81(1): 21-50.
- Carafoli E (1994) Biogenesis: plasma membrane calcium ATPase: 15 years of work on the purified enzyme. FASEB J 8(13): 993-1002.
- Clapham DE (2003) TRP channels as cellular sensors. Nature 4; 426(6966): 517-524.
- Vennekens R, Hoenderop JG, Prenen J, Stuiver M, Willems PH, (2000) Permeation and gating properties of the novel epithelial Ca(2+) cha4nnel. J Biol Chem 11; 275(6): 3963-3969.
- Vanden Abeele F, Roudbaraki M, Shuba Y, Skryma R, Prevarskaya (2003) N Store-operated Ca2+ current in prostate cancer epithelial cells. Role of endogenous Ca2+ transporter type 1. J Biol Chem 25; 278(17): 15381-15389.
- Chatzizacharias NA, Rosich Medina A, Dajani K, Harper S, Huguet E, et al. (2017) Surgical management of hepato-pancreatic metastasis from renal cell carcinoma. World J Gastrointest Oncol 15; 9(2): 70-77.
- Chiba S (2006) Notch signaling in stem cell systems. Stem Cell 24(11): 2437-2447.
- Cheng YY, Yu J, Wong YP, Man EP, To KF,et al. (2007) Frequent epigenetic inactivation of secreted frizzled-related protein 2 (SFRP2) by promoter methylation in human gastric cancer. Br J Cancer 8; 97(7): 895-901
- Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A et al. (2011) miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 219(1):211-217.
- Gutschner T, Hammerle M, Diederichs S (2013) MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl); 91(7): 791-801.
- Tian X, Xu G (2015) Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open 30; 5(9): e008653.
- Zhou X, Liu S, Cai G, Kong L, Zhang T, et al. (2015) Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci Rep Nov 2; 5:15972.
- Chen S, Ma P, Zhao Y, Li B, Jiang S, et al. (2017) Biological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: role of the MALAT-1-Livin protein interaction. J Physiol Sci 67(5): 577-585.
- Yang Y, Zhang J, Xia T, Li G, Tian T, et al. (2016) MicroRNA-210 promotes cancer angiogenesis by targeting fibroblast growth factor receptor-like 1 in hepatocellular carcinoma. Oncol Rep. Nov; 36(5): 2553-2562.
- Usachev YM, Toutenhoofd SL, Goellner GM, Strehler EE, Thayer SA (2001) Differentiation induces up-regulation of plasma membrane Ca(2+)-ATPase and concomitant increase in Ca(2+) efflux in human neuroblastoma cell line IMR-32. J Neurochem;76(6):1756-1765.
- Chan AS, Thorner PS, Squire JA, Zielenska M (2002) Identification of a novel gene NCRMS on chromosome 12q21 with differential expression between rhabdomyosarcoma subtypes. Oncogene. 2; 21(19): 3029-3037.
- Delgado Coello B, Santiago Garcia J, Zarain Herzberg A, Mas Oliva J (2003) Plasma membrane Ca2+-ATPase mRNA expression in murine hepatocarcinoma and regenerating liver cells. Mol Cell Biochem 247; (1-2):177-184.
- Aung CS, Kruger WA, Poronnik P, Roberts Thomson SJ, Monteith GR (2007) Plasma membrane Ca2+-ATPase expression during colon cancer cell line differentiation. Biochem Biophys Res Commun 20; 355(4): 932-936.
- Wu Y, Miyamoto T, Li K, Nakagomi H, Sawada N,et al. (2011) Decreased expression of the epithelial Ca2+ channel TRPV5 and TRPV6 in human renal cell carcinoma associated with vitamin D receptor. J Urol 186(6): 2419-2425.
-
Yasemin Yuyucu Karabulut*, Funda Bozkurt, Oznur Bucak, Emre Cagatay Kose, Bahar Taşdelen , Ali Nebioglu, Hasan Erdal Doruk , Mehmet Emin Erdal. A Novel Gene and Comparison of Gene Expressions in Low- and High-Grade Renal Cell Carcinomas. Annals of Urology & Nephrology. 3(4): 2023. AUN.MS.ID.000566.
-
Gene expression, Renal cell carcinoma, PMCA1, Nephropathology, Kidney
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.