 Research Article
Research Article
Microbial Load in Five Demersal Marine Fin and Shell Fishes from Nigerian Coastal Waters
Adeboyejo Akintade O*, Clark OE Dwin and Fajana O Paul
Lagos State University, Nigeria
Adeboyejo Akintade O, Department of Fisheries, Lagos State University, Ojo, Lagos, Nigeria.
Received Date: January 22, 2021; Published Date: March 16, 2021
Abstract
This study was conducted to assess the microbial load in five demersal species from the Nigerian coastal water (FAO, Area 34) with the aim of determining the bio-integrity of these species; Parapenaeopsis atlantica, Penaeus monodon, Penaeus notialis, Portunus validus and Cynoglossus browni. Five hundred and six (506) specimens in all were sampled from 32 fishing trawlers owned by Olokun Pisces Limited (Lat N 60 28ˡ 24ˡˡ, Long E 30 22ˡ 50ˡˡ). Microbiological analysis showed that total plate count (TPC) was observed to be similarly contained in all the fishes under consideration. TPC, Staphylococcus aureus, Yeast and Mould contained in the five species were not statistically significantly different from one another (P<0.05). amongst other species studied, C. browni had the highest TPC at 300C to range from 5.7×102 – TNTC cfu/g and 2.0×102 – TNTC (too numerous to count) for Yeast and Mould. The corresponding values for other species were within acceptable limit by FAO/WHO standard, which implies that batch of C. browni was not safe for consumption. The Most Probable Number (MPN) for the demersal species had values < 3cfu/g while the Coliform values for all the species was <10 cfu/g. Escherichia coli, Vibrio cholera, Salmonella spp, Shighellaspp, Streptococcus faecalis, Listeria monocytogenes, Clostridium botulinumand Vibrio parahaemolyticus were analyzed but were not detected (ND).
Introduction
According to FAO (2011) demersal fishes (flounders, halibuts, soles, cods, hakes, haddocks and miscellaneous demersal) contributed 12 percent of the total catches in 2009 (10.9 million tons), compared with almost 26 percent in the 1950s and 1970s. Catches of crustaceans (crabs, lobsters, shrimps, prawns, krill, etc.) contributed 6 percent (5.4 million tons) in 2009, slightly lower than 7 percent in 2002. Molluscs (abalones, conchs, oysters, mussels, scallops, clams, squids, octopus, etc.) increased slightly from 6 percent in the 1950s and 1970s to 7 percent (6.2 million tons) in 2009. Shrimps is an important seafood commodity representing a global industry with a market valued at over US $ 20 billion annually accounting for 19% of international trade. The Nigerian multispecies demersal stocks are exploited with a wide variety of artisanal gears; set gillnets, beach seines, large, meshed shark drift nets, hooks on longlines and various traps. Fish may be contaminated at various stages of transport, handling and processing. This contamination may be related to the raw materials, personnel and processing tools such as forklifts through leakage, insect and pest harborage. Additionally, seafood contamination may be caused by food borne pathogens which are naturally present in aquatic environments, such as Vibrio Spp., or derived from sewage contaminated water such as Salmonella spp. Consumption of these contaminated fish may cause infection or intoxication to the consumers. Vibrio cholera is responsible for the third-highest number of shellfish related illnesses, after non cholera Vibrio sp and Norwalk viruses. Toxigenic (epidemic biotype) infections are associated with profuse, watery diarrhea whereas non-toxigenic infections result in septicemia and mild gastroenteritis. In contrast to vibrio spp., the incidence of salmonella infections due to seafood consumption is still low compared to Salmonellosis associated with other foods.
However, detection of Salmonella spp. in seafood cannot be skipped as it is responsible for most of the food borne diseases or gastroenteritis characterized by diarrhea, abdominal cramp, vomiting, nausea and fever. According to Centers for Disease Control and Prevention, Salmonella is the leading cause of bacterial food-borne illness causing approximately 1.4 million non-typhoidal illnesses, 15,000 hospitalizations and 400 deaths in the USA annually. Many of the bacterial pathogens of fishes are flagellated, but a few have no flagella for locomotion. Pseudomonas sp have been reported to cause localized lesions and septicemia in some fishes. Flexibacter columnaris causes cotton wool disease (gill disease), Mycobacterium sp affects the fish leading to tuberculosis and Vibrio sp causes vibrio localized lesions and general disease in fish. Other important pathogens of fish are Aeromonas salmonicidia, A. Lychophila, Citrobacter freundi, Cytophagapsy chrophilia, Edward siellafada, Edward siellaretalin, Lactobacillus fisciola, Renibacterium salmoninarium, Streptococcus sp and Yeosinaructeri [1]. Though the broad distribution of the commercially exploited fish species groups is known, there is no adequate information on the composition of communities or on temporal and spatial distribution of stocks. In the circumstances of a very long coastline (about 800km), it is almost certain that many species form more than one stock. The distribution of demersal and pelagic fishes in the marine waters of Nigeria indicates discrete ecological fish communities, each of which is fairly homogenous. However, there is also ecological and micro geographical heterogeneity of fish communities, whilst migration of species from the estuaries and creeks to the open shelf areas and vice versa is known to occur. The aim of this project is to assess the Bio-integrity of five sea-foods from the Nigerian Coastal waters (FAO Area 34) with respect to their Microbial load. The specific objective is to isolate and identify health hazard microorganisms of public health importance associated with the demersal fish species. The high association of potential human pathogens among demersal fin and shellfish species suggests that if fish is not handled properly, or if consumed undercooked or uncooked, may lead to various infections and diseases to susceptible individuals. Hence, there is need to know or assess the Bio-integrity and the public health significance of microorganisms.
Materials and Methods
Study area
The Nigerian Coastal waters which fall within Eastern Central Atlantic FAO Area 34. Nigeria is bordered to the North by the Republic of Niger and Chad, to the West by the Republic of Benin, to the East by the Republic of Cameroon and to the South by the Atlantic Ocean. Nigeria has a coastline of approximately 853km facing the Atlantic Ocean. This coastline lies between latitude 40101 to 60 201 N and longitude 20 451 to 80 351 E. The Nigerian Coastal line has eight states it shares borders with (Figure 1).
They are Lagos, Ogun, Ondo, Rivers, Delta, AkwaIbom, Bayelsa and Cross River States. A trawler voyage of about fifty (50) days to and from covers the area and also the eight coastal states. Deep frozen samples (at -200C) of Cynoglossus browni (Nigerian tongue sole), Parapenaeopsis atlantica (Brown Shrimp), Penaeus notialis (White Shrimps), Penaeus monodon (Black tiger Shrimps) and Portunusvalidus (Smooth Swim Crab) were obtained from 32 fishing trawlers owned by a private fishing company. A total of 506 samples of each species for a period of 6 months collected were thawed. Sampling was done using bean trawls and other bottom trawls.
Sterilization of glass wares and working areas
Materials such as glass wares and media were sterilized before use. The glass wares including test tubes, pipettes, petri-dishes, and marcartney bottles were washed with detergent, rinsed thoroughly with clean tap water and allowed to air-dry before sterilizing in the hot air oven at 160°C for an hour. The media were sterilized and bench working areas were swabbed with cotton wool soaked in ethanol to sterilize before any microbiological analysis was carried out to avoid contamination.
Preparation of media
The media used in the experiment were weighed out according to the manufacturer’s specifications; this includes nutrient agar, Macconkey agar and blood agar. The media were properly weighed and distributed in conical flask containing distilled water and boiled gently to complete dissolution. The conical flask were plugged with cotton wool, covered with aluminum foil and sterilized in the autoclave at 121°C for 15 minutes at 1.06 mmHg. Agar media were allowed to cool to 45°C before dispensing aseptically into sterile petri dishes.
Preparation of the serial dilution
1ml of the original stock solution was aseptically poured into 9ml sterile distilled water and mixed thoroughly to give 10-1 dilution of the original Stock culture. 1ml of the dilution 10-1 of the original stock culture was aseptically poured into another tube of 9ml sterile distilled water to give 10ml of 10-2 dilution of the original stock culture. Repeating the above procedure, dilutions of 10-3, 10-4, 10-5, 10-6, 10-7, 10-8, 10-9, and 10-10 were obtained. The dilutions 10-2, 10-5, 10-8 and 10-10 were used.
Viable bacteria count on nutrient agar
The pour plate technique was employed. 0.5ml of dilution 10-1 of the stock culture was introduced into each of three sterilized nutrient agar plates. Sterile molten nutrient agar at about 4.5°C was added and then, mixed thoroughly and allowed to set undisturbed. The set agar plates were incubated at 37°C for 24 hours. This procedure was repeated using dilutions 10-2, 10-5, 10-8 and 10-10. Finally, the number of colonies per plate were counted and recorded.
Isolation of microorganisms from stock culture
A loopful each of the stock culture was inoculated into a sterile nutrient agar plate. The nutrients agar plate was incubated at 37°C for 24 hours for microbial growth.
Purification of bacteria
Characteristics colonies from original culture on nutrients agar plates were picked by sterile platinum wire loop and streaked to isolate on sterile nutrient agar, MacConkey agar and blood agar. These plates were incubated at 37°C for 24 hours for microbial growth. Distinct colonies were re-inoculated on to fresh sterile nutrient agar, MacConkey agar and blood agar plates for purity. Distinct colonies were Gram stained to know the Gram reaction and recorded. These isolates were transferred to their appropriate agar slants, labeled and incubated at 37°C for 24 hours for growth after which they were kept in the refrigerator at 4°C for identification.
Gram’s staining
A very small loop of distilled water was placed on a clean slide the inoculating wire loop was sterilized by flaming it until it was red hot in the blue flame of a Bunsen burner. The loop was allowed to cool and a small portion of the colony of the organisms to be stained was picked and smeared in the drop of distilled water on the slide and spread into a thin smear along the slide. The smear was allowed to air dry and heat fixed briefly over flame. The smear was stained with 1% crystal violet for 60 seconds and washed with distilled water. The slide was decolorized rapidly with 75% alcohol and washed off immediately with distilled water. The slide was finally flooded with counter stain-saframine for 30 seconds and washed off with distilled water and air dried. This was then observed under the microscope with the aid of oil immersion lens (108 objective lens).
Motility test
The organisms were grown in MRS broth for 48 hours at 30°C. A drop of MRS broth was put on a stainless slide and examined under the oil immersion lens of the microscope. The motile cells were seen to move about randomly. Formation of a very deep purple colouration within 10 seconds indicated a positive reaction while absence of deep colouration indicated a negative reaction.
Biochemical test
Oxidase test: A piece of filter paper was placed into a clean petri dish, 2 or 3 drops of freshly prepared oxidase reagent was added. A small portion of the organism to be identified was smeared on the filter paper, using a glass rod (not an oxidising wire loop). A blue purple colour indicates positive result (WHO, 1989).
Catalase test: A drop of 30% hydrogen peroxide was placed on to the centre of a slide and sterile wire loop was used to pick small portion of the microorganisms to be identified from nutrient agar plate into the hydrogen peroxide for immediate gas bubble formation or foaming (immediately) indicates positive result (WHO, 1989).
Coagulase test: A drop of physiological saline was placed on two separate slides. A colony of the test organism was emulsified in each of the drops to make suspensions. A drop of plasma was then added and mixed gently with the suspensions. Clumping of the organisms in 10 seconds indicate positive result (WHO, 1989).
Citrate utilization test: A slant of a citrate agar was aseptically inoculated with organism to be identified using a sterile wire loop. The inoculated citrate agar slant was incubated at 37°C for 24 hours and observed for colour change daily for up to 4 days. Turbidity and blue colour indicate positive test (WHO, 1989).
Urease test: A slant of urea agar was aseptically inoculated in then organism to be identified using a sterile wire loop and incubated at 37°C for 24 hours and observed for colour change. A red pink colour indicates positive result (WHO, 1989).
Hydrogen sulphide production: Filter paper was cut into slip of about 5 - 10 mm wide and 50 - 60 mm long. These were impregnated with hot saturated aqueous lead acetate solution dried at 50°C - 60°C and stored in tightly closed container before used. The lead acetate paper was then inserted between the cap or the plug of the tube containing the organism in nutrient broth or peptone water. This was examined for seven days. A brown or black colour indicates positive result (WHO, 1989).
Indole reaction: The microorganism to be identified was inoculated into trytophane broth for 48 hours at 37 °C, 5 drops of Kovac’s reagent was then added. A deep red colour indicates positive result (WHO, 1989).
Gelatin hydrolysis: Stab cultures of the organisms to be identified was inoculated into a solidified column of meat petone gelatin and incubated at room temperature for three to six days. The hydrolysis of gelatin may take place in the form of a nail, funnel cup, bubble or in layers. If the enzyme was absent, the organism grows in the surface (WHO, 1989).
Voges proskauer: This was performed to confirm the production of acetylmethylcarbinol (Acetoin) or 2, 3 - butane diol from acid produced from glucose using 10ml of the 3-day old cultures of each isolate in glucose broth was added, 0.5ml of 8% alcoholic solutions of x-naphtol and 0.5ml of 16% KOH solution. The tubes were shaken at intervals to ensure maximum aeration. The development of pink colour in 2 - 5 minutes becoming crimson in 30 minutes or red colour within 5 - 10 minutes indicates positive result (WHO, 1989).
Methyl red test: Many bacteria produce organic acids from the metabolism of glucose. The organic acids produced lower the pH of the growth medium to less than pH 4.4; this changed the colour of the methyl red indicator to its red state. Ten millilitres of glucose phosphate broth was dispensed into screw caps tubes and sterilised. Inoculating with test organisms was subsequently done and the set up incubated at 30°C. A resultant definite red colouration was considered positive.
Starch hydrolysis :7g of nutrient agar was weighed and put into a conical flask containing 250ml distilled water. 0.7g of starch was weighed and added to the solution and it was thoroughly mixed together. This was the sterilised at 121°C for 15 minutes after which it was poured into sterile plates and allowed to solidify. The organism to be tested was then streaked on it using a sterile platinum wire loop. This was then incubated at 37°C for 24 hours. Drops of iodine solution were added and a blue-black colouration gave a positive resul
Sugar fermentation: This is the ability of the isolates to utilize carbon sugar as the only source of energy. The sugar fermentation test was carried out using the method of Nair [3].1% solution (1g of the sugar under test in 100ml of distilled water was prepared separately in Maccartney bottles. 1ml of 1% solution was added to 9ml of phenol red peptone water in test tubes and mixed gently. The test tubes were covered with cotton wool and sterilised at 121°C for 15 minutes at 1.06mmHg. On cooling, the tubes were inoculated with bacteria isolate and incubated at 37°C for 48 hours. Colour change from red to yellow indicates acid production while formation of gas in Durham tube indicates gas production (WHO, 1989).
Result
The Most Probable Number (MPN) for the demersal species had values < 3 while the Coliform values for all the species was <10. The total count plate (TPC) values for P. validus, P. notialis, P. monodon, P. atlantica and C. browni at 300C was 7.58±0.65, 6.44± 0.26, 7.37 ± 0.73, 7.23± 0.38 and 8.87 ± 1.37 respectively. Staphylococcus aureus values for P. validus, P. notialis, P. monodon, P. atlantica and C. browni was 2.38 ± 0.36, 3.75 ± 0.54, 3.50 ± 0.55, 3.02 ± 0.37 and 3.89 ± 0.31 respectively. Yeast and Mould values for P. validus, P. notialis, P. monodon, P. atlantica and C. browni was 5.45 ± 0.64, 5.67 ± 0.50, 4.85 ± 0.38, 5.33 ± 0.37 and 5.99 ± 0.27 respectively (Table 1).
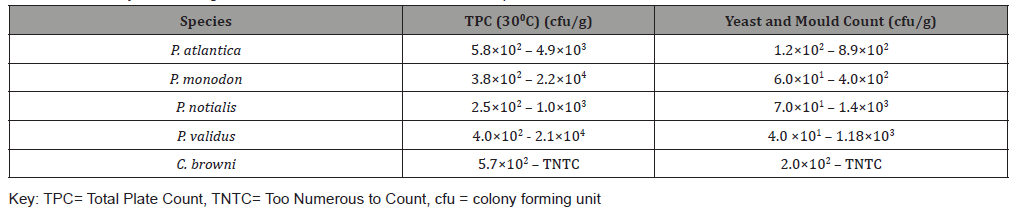
Table 2 shows summary of the range of variables for Total plate count and Yeast and Mould count. P. atlantica had viable count range from 5.8×102 – 4.9×103 for TPC and 1.2×102 – 8.9×102 for Yeast and Mould. P. monodon had viable count range from 3.8×102 – 2.2×104 for TPC and 6.0×101 – 4.0×102 for Yeast and Mould. P. notialis had viable count range from 2.5×102 – 1.0×103 for TPC and 7.0×101 – 1.4×103 for Yeast and Mould. P. validus had viable count range from 4.0×102 - 2.1×104 for TPC and 4.0 ×101 – 1.18×103 for Yeast and Mould while C. browni had viable count range from 5.7×102 – TNTC (Too numerous to count) for TPC and 2.0×102 – TNTC for Yeast and Mould (Table 2).
Table 1:Summary of Microbial load for the fish species.
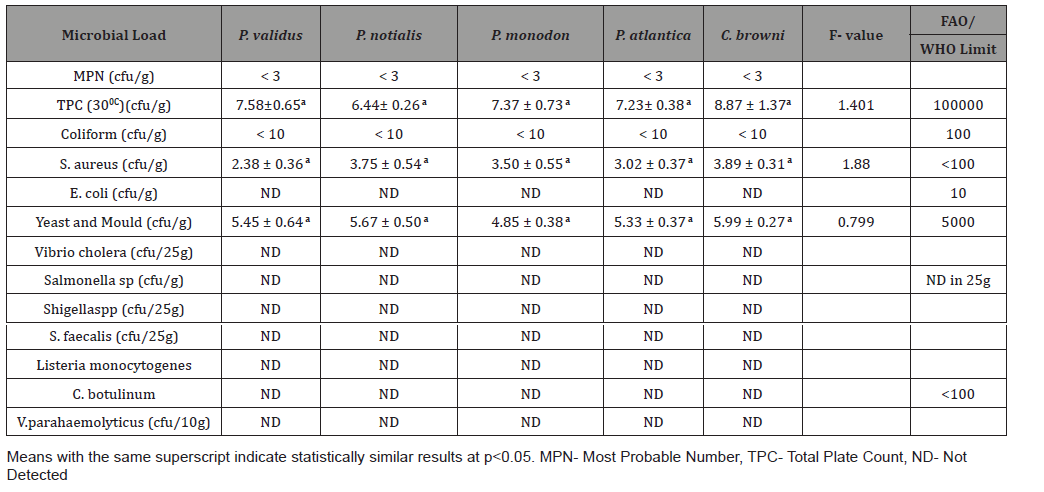
Table 2:Summary of the Range of Microbial count of the five demersal Species.
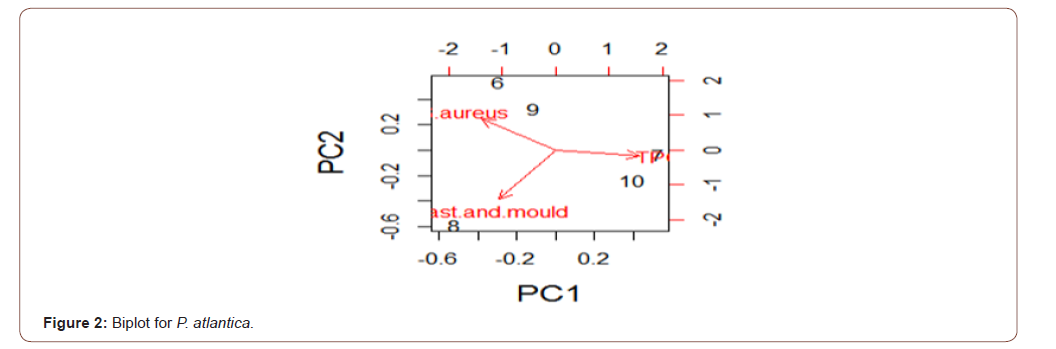
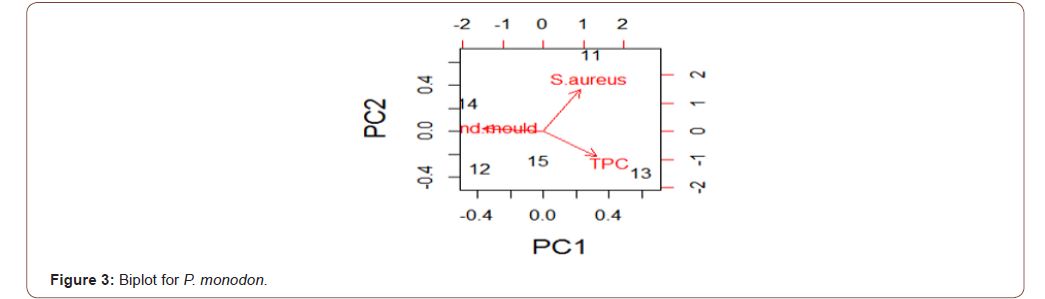
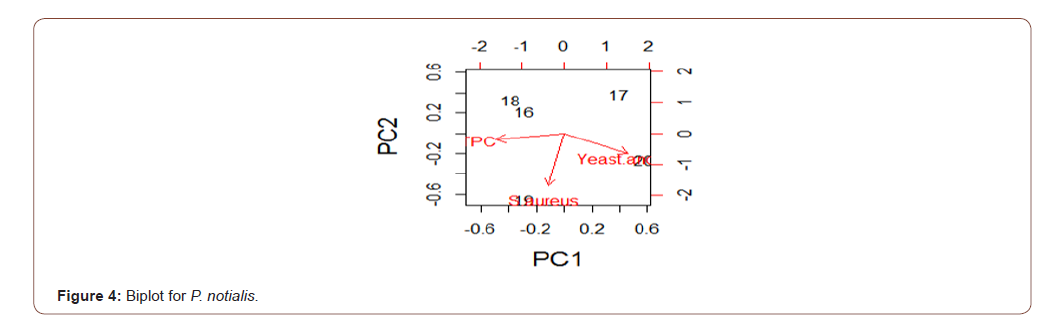
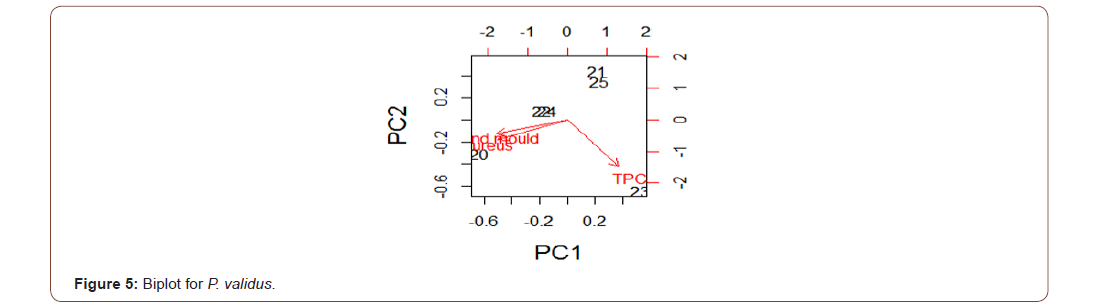
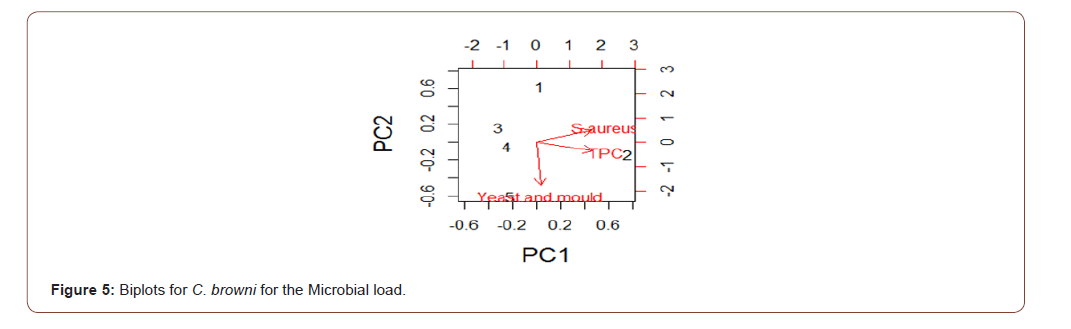
Analysis of variance for the microbial load for the five species in natural logarithm values for the Total Plate Count (TPC), Staphylococcus aureus, Yeast and Mould as shown in Table 1, TPC was observed to be similarly contained in all the species under consideration. It was also observed, at 95% confidence level, that TPC, Staphylococcus aureus, Yeast and Mould contained in the five species were not statistically significantly different from one another. Escherichia coli, Vibrio cholera, Salmonella sp, Shighella sp, Streptococcus faecalis, Listeria monocytogenes, Clostridium botulinumand Vibrio parahaemolyticus were analyzed but not detected (ND). The biplots for the microbial load for the five species considered are shown in Figures 2-6. The biplot for P. atlantica (Figure 2, 3) were observed to contain TPC similarly while samples 4 and 5 were observed to contain more S. aureus than 6, which was observed to contain Yeast and Mould the most compared to others. Figure 3 contained the most S. aureus while fin fish contained more TPC compared with others for P. monodon. Figure 4 contained more Yeast and Mould compared with others (Figures 2-6).
Discussion and Conclusion
This study focused on isolating and identifying Microorganisms in the edible part of the species (shrimps, crab and sole fish) that are of public health importance especially in the food industry. Tissue samples for microbiological analysis were taken at a temperature of -200C. Vibrio spp and E.coli was not detected in this study unlike Chuma et al [2] who took samples for bacterial isolation at different storage temperatures such as 280C, 40C and -50C and detected both organisms in the external surface, tissue , gills and intestines in his study of the Quality assessment of Croaker (Pseudotolithus elongatus), Total Plate Count (TPC) was observed to be similarly contained in all the demersal species under consideration. It was also observed, at 95% confidence level, that Staphylococcus aureus and Yeast & mould contained in the five demersal species were not statistically significantly different from one another. The highest mean weight of the three species reported by Akinyemi & Fajana [1] was found in Chrysichthys nigrodigitatus to be 77.38g ± 7.615 and highest viable bacterial count to be 5.8×104 – 8.00×104cfu/ml in the skin of Distichodusen gycephalus while this study revealed the highest mean weight amongst the five demersal species to be 411.09g ± 15.27 in Cynoglossus browni and the highest Total Plate Count (TPC) at 300C to be 5.7×102 – TNTC cfu/g (Table 2) in Cynoglossus browni. The TPC for P. atlantica, P. monodon, P. notialis and P. validus were within accepted limit by FAO/WHO standard except for a sample of C. browni that had TNTC value for TPC and for Yeast and Mould which implies that batch of C. browni was not safe for consumption. All other samples of C. browni had TPC within acceptable limits by FAO/WHO. E. coli, Salmonella sp, Listeria monocytogenes, Vibrio parahaemolyticus, Streptococcus faecalis, Clostridium botulinum, Shigella sp and Vibrio cholera were tested and not detected (ND) in the selected samples.
The MPN (Most Probable Number) is a method used to estimate the concentration of viable microorganisms in a sample by means of replicate liquid broth growth in ten-fold dilutions. It is commonly used in estimating microbial population and is particularly useful with samples that contain particulate material that interferes with plate count enumeration methods. The presence of very few fecal Coliform bacteria would indicate that water probably contains no disease causing organisms, while the presence of large numbers of fecal coliform bacteria would indicate a very high probability that the water could contain disease-producing organisms making the water unsafe for consumption. However, the MPN values for all microorganisms count or remunerated in all the samples used for this study were <3. Some of the low values of microorganisms gotten could be as a result of strict instruction and briefing the Quality assurance unit of the company passed to the sailors especially not fishing in muddy area, polluted water and shallow waters so as to reduce contamination.
The weight of the species increased more in proportion or less to the cube of its length as depicted by the slope (b). Portunus validus has the highest growth in weight relatively to the other species with a condition factor (K) of 8.77. C. browni had the highest Total Plate Count (TPC) at 30 0C to be 5.7×102 – TNTC cfu/g and 2.0×102 – TNTC (too numerous to count) for Yeast and Mould. The corresponding values for other species were within acceptable limit by FAO/WHO standard, which implies that batch of C. browni that had TPC to be TNTC was not safe for consumption, which compromises its Bio-integrity. Microorganisms such as Coliform and Staphylococcus aureus that was detected were of public health importance especially in the food industry. Escherichia coli, Vibrio cholera, Salmonella sp, Shighella sp, Streptococcus faecalis, Listeria monocytogenes, Clostridium botulinumand Vibrio parahaemolyticus were analyzed and results showed not detected (ND).
The Nigerian coastal water is a very large area that shares border with eight States. Further studies should be investigated as it will broaden the knowledge of environmental influence on the bio-integrity of fish (fin fish, crustaceans, Molluscs etc.) found within this territory. Based on the study and its results, the microbial load presupposes that there are preponderance of heavy load of microorganisms of serious health importance, the relevant authority need to enforce proper handling of sea-foods during processing and preservation.
Acknowledgment
None.
Conflict of Interest
No conflict of interest.
References
- Akinyemi, Fajana (2011) Antibiotics sensitivity and Occurrence of bacteria Originated from the Skin, Gills and Buccal cavity of Chrysichthys nigrodigitatus, Distichodus engycephalus and Synodontis schall sampled in Arakanga Reservoir, Abeokuta, Ogun State, Nigeria. J Fisheries Sciences.com pp.164-171.
- Chuma C Okoro, Olusimbo O Aboaba, Ola J Babajide (2010) Quality Assessment of a common Nigerian Marine Fish, Croaker (Pseudotolithus elongatus) under different Storage conditions. New York Science Journal pp.29-36.
- Nair PS, Surendren P K (2005) Biochemical characterization of Latic acid bacteria isolated from fish and prawn. Journal of culture collections 4: 48-52.
- Ajayi TO, Talabi SO (1984) The potential and strategies for optimum utilization of the fisheries resources of Nigeria. NIOMR Technical Paper No 18: 25.
- Benjamin Carvalho Pinto, Francisco Gerson Araujo (2007) Assessing of biotic integrity of the fish community in a heavily impacted segment of a tropical river in Brazil. Braz arch biol Technol 50(3).
- Nwosu FM, Ita EO, Enin UI (2011) Fisheries management in Nigeria: A case study of the marine fisheries policy. International Research Journal of Agricultural Science and Soil Science 1(3): 070 - 076.
- Omolere EA (2012) Marine fisherman about 60 years at Ogheye. Oral Interview, p.12.
- Sara Safaeian, Saeid Khanzadi (2018) Microbiology of Fish and Seafood. National Conference on Recent Advances in Engineering and Modern Sciences. IRIB International Conference Center.
- Sea Around Us (2007) A global database on marine fisheries and ecosystem.
- Wikipedia (2019) Biological Integrity.
-
Adeboyejo Akintade O, Clark OE Dwin, Fajana O Paul. Microbial Load in Five Demersal Marine Fin and Shell Fishes from Nigerian Coastal Waters. Ad Oceanogr & Marine Biol. 2(4): 2021. AOMB.MS.ID.000543.
-
Parapenaeopsis atlantica, Penaeus monodon, Penaeus notialis, Portunus validus, Cynoglossus browni, Staphylococcus aureus, Microbial load
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






