 Mini Review
Mini Review
LRRK2 Signalling Pathways in Parkinson’s Disease
Jinwei Zhang*
Institute of Biomedical and Clinical Sciences, College of Medicine and Health, University of Exeter, Hatherly Laboratories, Exeter, EX4 4PS, UK
Jinwei Zhang, Institute of Biomedical and Clinical Sciences, College of Medicine and Health, University of Exeter, Hatherly Laboratories, Exeter, EX4 4PS, UK.
Received Date:January 08, 2019; Published Date: January 16, 2019
Abstract
Parkinson’s disease (PD) is a common neurodegenerative disorder with the pathological hallmark of progressive loss of dopamine neurons in affected brains. There is no cure for PD and current therapy does not halt the underlying degenerative process. The pathogenesis of PD is not fully understood but is likely caused by a combination of genetic and environmental factors. Several genes are associated with the onset and progression of familial PD. Mutations in LRRK2 are the most frequent known cause of lateonset PD. Many studies have been conducted to elucidate the functions of LRRK2 and identify effective LRRK2 inhibitors for PD treatment. In this review, I discuss the role of LRRK2 in PD and recent progress in studying LRRK2 cellular functions and the use of LRRK2 inhibitors as therapeutic agents.
Introduction
Parkinson’s disease (PD) was first described by Dr. James Parkinson in 1817 as a “shaking palsy” [1] is the second most common neurodegenerative disorder after Alzheimer’s disease (AD). PD affected 6.2 million people and resulted in about 117,400 deaths globally in 2015 [3], typically occurs in people over the age of 60, of which about one percent are affected [3], with an incidence of 1.7% in individuals aged over 65 years [4]. PD is characterized by the selective loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc) of the midbrain in affected brains. There is no cure for PD and current therapy does not halt the underlying degenerative process, although several clinical approaches, including levodopa treatment and deep brain stimulation, are currently used to manage the symptoms of PD.
Therefore, it is vitally important to elucidate the mechanisms underlying PD pathogenesis, to predict disease onset and provide scientific guidance for targeted therapy. Recent genetic studies have revealed an underlying genetic cause in at least 10% of all PDcases [5] which provides new opportunities for discovery of molecularly targeted therapeutics that may ameliorate neurodegeneration. Among the genes associated with PD, leucine-rich repeat kinase 2 (LRRK2) has emerged as the most relevant one to PD pathogenies, since its mutations were identified in PD patients in 2004 [6,7]. More than 40 mutations of LRRK2 have been found in both familial and sporadic forms of PD [8,9], among which, a missense mutation, G2019S, is frequently found in not only familial but also sporadic Parkinson’s disease cases [10].
The G2019S mutation enhances kinase activity, suggesting that small molecule LRRK2 kinase inhibitors may be able to block aberrant LRRK2-dependent signaling in Parkinson’s disease [11,12]. In this review, I discuss the role of LRRK2 in PD pathogenesis and recent progress in the development of LRRK2 inhibitors for PD treatment.
LRRK2 Biology
LRRK2 is a large gene whose transcript encodes a 2527 amino acid protein (286 kDa) that is comprised of 51 exons. Sequence analysis predicts that LRRK2 contains multiple domains, including an armadillo repeats (ARM), an ankyrin-like (ANK) domain, leucine-rich repeat (LRR) domain, a Roco family of G-proteins comprising a Ras-of-complex (Roc) domain followed by its associated C-terminal of Roc (COR) domain, a mitogen-activated protein kinase (MAPK) domain and a C-terminal WD40 domain (Figure 1). The presence of both protein interaction domains (ARM, ANK, LRR and WD40) and the enzymatic domains (ROC and MAPK) within LRRK2 suggests that this protein may serve as a scaffold for assembly of a multiprotein complex and act as a central integrator of multiple signaling pathways. The multiple allosteric and enzymatic functions within one protein make LRRK2 an excellent therapeutic target. Several proteins, including ezrin/ radixin/moesin[13], 4E-BP[14], MKKs [15,16], β- tubulin [17], α-synuclein[18], peroxiredoxin 3[19], Akt1[20] and ArfGAP1 [21,22], have been reported to be phosphorylated by LRRK2, and validated as non-physiological substrates of LRRK2. Recently, progress in studying cellular functions of LRRK2 has been made, that Ser1292 autophosphorylation occurs in vivo and is enhanced by several LRRK2 mutations and could be inhibited brainpenetrating LRRK2 kinase inhibitor [23]. Further, the role of LRRK2 in vesicular membranes could be mediated by the ability of LRRK2 to phosphorylate a subgroup of Rab GTPases, including including RAB8A (Thr72) and RAB10 (Thr73), at highly conserved sites located in the center of the effector binding motif [24,25]. However, the exact interplay between LRRK2 and Rab GTPases remains unclear. More elegant studies on the functional substrates of the LRRK2 kinase domain and their downstream signalling pathways are needed to understand the function of LRRK2 and its role in PD (Figure 1).
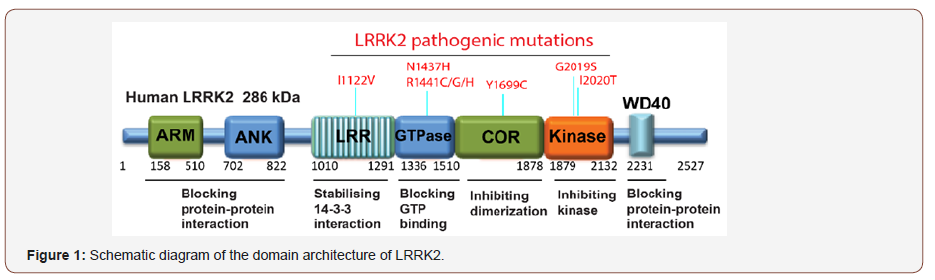
A linear representation of LRRK2 sequence and the domain organization with some of the most commonly occurring Parkinson’s disease mutations (including p. I1122V, p.N1437H, p.R1441C/G/H, p.Y1699C, p.G2019S, and p.I2020T) annotated on these domains. The regions and associated functions of the LRRK2 gene are also indicated.
LRRK2 as a Therapeutic Target
Protein kinases have become one of the most important classes of drug targets in medicine, particularly in the field of oncology [26]. In the past decade, more than 20 different drugs targeting kinases have been approved for clinical use in humans for the treatment of various types of cancer [27]. LRRK2 kinase activity is critically linked to clinical effects, and the most prevalent PD mutation, LRRK2 G2019S in the kinase domain, enhances kinase activity by 2-4 folds [28], LRRK2 kinase inhibitors are therefore being actively pursued. Some non-selective inhibitors including staurosporin, K252A and Su-11248 (Sunitinib) were found to inhibit LRRK2 with their IC50 values in nanomolar range [29,30]. ROCK inhibitors such as isoquinolinesulfonamides hydroxy fasudil and H1152, and the structurally unrelated Y-27632, have also been found to inhibit LRRK2 with similar efficiencies (low micromolar range) [30]. Recently, first-generation ‘tool’ inhibitors that exhibit excellent potency and selectivity for LRRK2 such as LRRK2-IN-1 [31], GSK2578215A [32], TAE684 and CZC-251469 [33] have been reported with nanaomolar range, but incapable of crossing the blood brain barrier. Soon after, HG-10-102-01 [34], GNE-7915 [35], JH-II-127 [36], PF-06447475[37], MLi-2 [38], 5-substituted- N-pyridazinylbenzamide derivatives [39] and 5-azaindazole inhibitors [40] are reposted as selective and brain penetrant LRRK2 kinase inhibitors.
LRRK2 is widely expressed in human tissues. Loss of LRRK2 has been shown to have a pathological phenotype in kidney and lung tissues [41], pharmacological LRRK2 kinase inhibition induces LRRK2 protein destabilization and proteasomal degradation [42], suggesting that normal LRRK2 kinase activity is needed for normal physiological function and LRRK2 kinase inhibitors may have severe adverse effects on normal tissues. However, it would be novel if a LRRK2 kinase inhibitor could be developed for specifically targeting LRRK2 active form [G2019S] without affecting the LRRK2 wild-type kinase. Indeed, clinical success of inhibition of kinases harboring disease causing driver mutations has been quite dramatic in the field of oncology: BRAF(V600E) in melanoma [42], mutant EGFR [43] and EML4-ALK in lung cancer [44], and BCRABL in chronic myelogenous leukemia [45]. Therefore it would be possible to pharmacologically interrogate the effects of inhibition of LRRK2[G2019S] in PD to develop appropriate tool compounds using structure based design and evaluate them in available PD disease models.
Acknowledgemnet
None.
Conflict of Interest
No conflict of interest.
References
- Shulman JM, De Jager PL, Feany MB (2011) Parkinson’s Disease: Genetics and Pathogenesis. Annu Rev Pathol 6(6): 193-222.
- GBD 2015 Disease and Injury Incidence and Prevalence Collaborators (2016) Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388(1053): 1545-1602.
- Rektorová I, Balá M, Rusina R, Matìj R (2016) Parkinson’s Dementia, Lewy Body Disease and Other Synucleinopathies. In: Lisak RP, Truong DD, Carroll WM, Bhidayasiri R(eds.), International Neurology, Second Edition: 776.
- Zhang ZX, Roman GC, Hong Z, Wu CB, Qu QM, et al. (2005) Parkinson’s disease in China: prevalence in Beijing, Xian, and Shanghai. Lancet 365(9459): 595-597.
- Daniels V, Baekelandt V, Taymans JM (2011) On the road to leucine-rich repeat kinase 2 signalling: evidence from cellular and in vivo studies. Neurosignals 19(1): 1-15.
- Paisan Ruiz C, Jain S, Evans EW, Gilks WP, Simón J, et al. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44(4): 595-600.
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, et al. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44(4): 601-607.
- Berg D, Schweitzer KJ, Leitner P, Zimprich A, Lichtner P, et al. (2005) Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson’s disease*. Brain 128: 3000-3011.
- Khan NL, Jain S, Lynch JM, Pavese N, Abou-Sleiman P, et al. (2005) Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson’s disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain 128: 2786-2796.
- Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, et al. (2008) Phenotype, genotype, and worldwide genetic penetrance of LRRK2- associated Parkinson’s disease: a case-control study. Lancet Neurol 7(7): 583-590.
- Greggio E, Cookson MR (2009) Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro 1(1): e00002.
- Kumar A, Cookson MR (2011) Role of LRRK2 kinase dysfunction in Parkinson disease. Expert Rev Mol Med 13: e20.
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, et al. (2009) Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci 29(44): 13971-13980.
- Imai Y, Stephan Gehrke, Hua-Qin Wang, Ryosuke Takahashi, Kazuko Hasegawa, et al. (2008) Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 27(18): 2432-2443.
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M (2009) The Parkinson disease- associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem 109(4): 959-968.
- Hsu CH, Chan D, Greggio E, Saha S, Guillily MD, et al. (2010) MKK6 binds and regulates expression of Parkinson’s disease-related protein LRRK2. J Neurochem 112(6): 1593-1604.
- Gillardon F (2009) Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability a point of convergence in parkinsonian neurodegeneration? J Neurochem 110(5): 1514-1522.
- Qing H, Wong W, McGeer EG, McGeer PL (2009) Lrrk2 phosphorylates alpha synuclein at serine 129: Parkinson disease implications. Biochem Biophys Res Commun 387(1): 149-152.
- Angeles DC, Gan BH, Onstead L, Zhao Y, Lim KL, et al. (2011) Mutations in LRRK2 increase phosphorylation of peroxiredoxin exacerbating oxidative stress-induced neuronal death. Hum Mutat 32: 1390-1397.
- Ohta E, Kawakami F, Kubo M, Obata F (2011) LRRK2 directly phosphorylates Akt1 as a possible physiological substrate: impairment of the kinase activity by Parkinson’s disease-associated mutations. FEBS Lett 585(14): 2165-2170.
- Stafa K, Trancikova A, Webber PJ, Glauser L, West AB, et al. (2012) GTPase activity and neuronal toxicity of Parkinson’s disease-associated LRRK2 is regulated by ArfGAP1. PLoS Genet 8(2): e1002526.
- Xiong Y, Yuan C, Chen R, Dawson TM, Dawson VL (2012) ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J Neurosci 32(11): 3877-3886.
- Sheng Z, Zhang S, Bustos D, Kleinheinz T, Le Pichon CE, et al. (2012) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med 4(164): 164ra161.
- Steger M, Tonelli F, Ito G, Davies P, Trost M, et al. (2016) Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 5: e12813.
- Steger M, Diez F, Dhekne HS, Lis P, Nirujogi RS, et al. (2017) Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. Elife 6: e31012.
- Zhang J, Yang PL, Gray NS (2009) Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 9(1): 28-39.
- Luzon Toro B, Rubio de la Torre E, Delgado A, Perez Tur J, Hilfiker S (2007) Mechanistic insight into the dominant mode of the Parkinson’s disease-associated G2019S LRRK2 mutation. Hum Mol Genet 16(17): 2031-2039.
- Anand VS, Reichling LJ, Lipinski K, Stochaj W, Duan W, et al. (2009) Investigation of leucine-rich repeat kinase 2: enzymological properties and novel assays. FEBS J 276(2): 466-478.
- Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, et al. (2009) Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson’s disease. Biochem J 424(1): 47-60.
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, et al. (2011) Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol 7(4): 203-205.
- Reith AD, Bamborough P, Jandu K, Andreotti D, Mensah L, et al. (2012) GSK2578215A; a potent and highly selective 2-arylmethyloxy-5- substitutent-N-arylbenzamide LRRK2 kinase inhibitor. Bioorg Med Chem Lett 22: 5625-5629.
- Zhang J, Deng X, Choi HG, Alessi DR, Gray NS (2012) Characterization of TAE684 as a potent LRRK2 kinase inhibitor. Bioorg Med Chem Lett 22(5): 1864-1869.
- Choi HG, Jinwei Zhang, Xianming Deng, John M. Hatcher, Matthew P Patricelli, et al. (2012) Brain Penetrant LRRK2 Inhibitor. ACS Med Chem Lett 3: 658-662.
- Estrada AA, Liu X, Baker-Glenn C, Beresford A, Burdick DJ, et al. (2012) Discovery of highly potent, selective, and brain-penetrable leucine-rich repeat kinase 2 (LRRK2) small molecule inhibitors. J Med Chem 55(22): 9416-9433.
- Hatcher JM, Zhang J, Choi HG, Ito G, Alessi DR, et al. (2015) Discovery of a Pyrrolopyrimidine (JH-II-127), a Highly Potent,Selective, and Brain Penetrant LRRK2 Inhibitor. ACS Med Chem Lett 6(5): 584-589.
- Henderson JL, Kormos BL, Hayward MM, Coffman KJ, Jasti J, et al. (2015) Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)- 7H-pyrrolo[2,3-d] pyrimidin-5-yl] benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor. J Med Chem 58(1): 419-432.
- Fell MJ, Mirescu C, Basu K, Cheewatrakoolpong B, DeMong DE, et al. (2015) MLi-2, a Potent, Selective, and Centrally Active Compound for Exploring the Therapeutic Potential and Safety of LRRK2 Kinase Inhibition. J Pharmacol Exp Ther 355: 397-409.
- Ding X, Stasi LP, Dai X, Long K, Peng C, et al. (2019) 5-Substituted- N-pyridazinylbenzamides as potent and selective LRRK2 inhibitors: Improved brain unbound fraction enables efficacy. Bioorg Med Chem Lett 29(2): 212-215.
- Osborne J, Birchall K, Tsagris DJ, Lewis SJ, Smiljanic-Hurley E, et al. (2018) Discovery of potent and selective 5-azaindazole inhibitors of leucine- rich repeat kinase 2 (LRRK2) - Part 1. Bioorg Med Chem Lett: S0960-894X(18)30934-X.
- Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, et al. (2012) Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener 7: 2.
- Lobbestael E, Civiero L, De Wit T, Taymans JM, Greggio E, et al. (2016) Pharmacological LRRK2 kinase inhibition induces LRRK2 protein destabilization and proteasomal degradation. Sci Rep 6: 33897.
- Janne PA, Suresh S Ramalingam, James Chih Hsin Yang , Myung Ju Ahn, Dong Wan Kim, et al. (2014) Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients (pts) with EGFR inhibitor-resistant non-small cell lung cancer (NSCLC). Journal of Clinical Oncology 32.
- Sullivan I, Planchard D (2016) ALK inhibitors in non-small cell lung cancer: the latest evidence and developments. Ther Adv Med Oncol 8(1): 32-47.
- Schenone S, Brullo C, Botta M (2010) New Opportunities to Treat the T315I-Bcr-Abl Mutant in Chronic Myeloid Leukaemia: Tyrosine Kinase Inhibitors and Molecules that Act by Alternative Mechanisms. Current Medicinal Chem 17(13): 1220-1245.
-
Jinwei Zhang. LRRK2 Signalling Pathways in Parkinson’s Disease: A Mini Review. Arch Neurol & Neurosci. 2(3): 2019. ANN. MS.ID.000540.
-
LRRK2 signalling, Parkinson's disease, Neurodegenerative disorder, Brain, Pathogenesis, Alzheimer’s disease, Biology, Enzymatic functions, Lung tissues, Pharmacological LRRK2 kinase inhibition, Kinases harboring disease
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






