 Research Article
Research Article
An Algorithm-Based Computational Method for Overcoming Worldwide Antimicrobial Resistance Exacerbation in COVID-19 Pandemic Era
Hyunjo Kim1* and Jae-Hoon Song2
1School of Pharmacy, Yonsei University, South Korea
2Chairman of Asian-Pacific Research Foundation for Infectious and ANSORP, South Korea
Hyunjo Kim, School of Pharmacy, Yonsei University, South Korea
Received Date: October 09, 2020; Published Date: October 28, 2020
Abstract
The use of antibiotics in COVID-19 pandemic era will inevitably exacerbate antimicrobial resistance (AMR) and could ultimately lead to more deaths and morbidity as an unintended consequence of this long-term pandemic. Although AMR was already spreading rapidly before the emergence of COVID-19, the impact of the pandemic may be more pronounced. The wider implications of the pandemic for health and social care systems must be characterized and these data must be gathered to inform and update national strategies that address the long-term challenges of AMR while preserving access to effective drugs. Thus, world needs such level of dedication to reverse the trajectory of AMR by ensuring that research continues to develop antimicrobial drug pipeline, sustainable investment in health systems, and strengthening of antimicrobial stewardship.
Keywords:Antibiotics, COVID-19 Pandemic, Antimicrobial Resistance (AMR), Multi-Drug Resistant (MDR), Health Systems, Antimicrobial Stewardship
Abbreviations: Coronavirus Disease-19 (COVID-19); Antimicrobial Resistance (AMR); Severe Acute Respiratory Syndrome coronavirus 2 (SARSCoV- 2); Multi-Drug Resistant (MDR); Chronic Obstructive Pulmonary Disease (COPD); Antimicrobial Stewardship (AMS); Angiotensin-Convert- ing Enzyme (ACE); Acute Respiratory Distress Syndrome (ARDS); Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV); Quantum Universal Exchange Language (Q-UEL).
Introduction
COVID-19 pandemic has changed the focus of healthcare around the globe and antibiotics may be indicated in patients with COVID-19 due to suspected or confirmed bacterial superinfection [1-3]. Many studies reporting on patients hospitalized with COVID-19 have been revealed widespread use of antimicrobial therapies as part of the package of clinical care in some countries [4-7]. Although antibiotic treatment of secondary bacterial pneumonia associated with influenza is accepted clinical practice, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is not influenza. In patients with COVID-19 who have co-infections, antibiotic or antiviral treatment is appropriate but those patients may be in a minority. Before the SARS-CoV-2 pandemic, overcoming antimicrobial resistance (AMR) already demanded urgent global action. Now that we are in the midst of a pan- demic, understanding the pathogenesis of SARS-CoV-2 infection, and the potential for bacterial coinfections, is imperative. Until then, efforts to prioritize antibiotic stewardship around the world must be redoubled for potential long-term impact on antimicrobial resistance. The COVID-19 has placed huge strains on health and social care systems and resources globally. It is important to con- sider the short and longer-term consequences that COVID-19 may have on antimicrobial use and drug-resistant infections. A concerning potential consequence of the COVID-19 pandemic is the long-term propagation of AMR in the acute care setting, resulting from increased patient exposure to antimicrobials, often sub-optimally or inappropriately used [8,9]. However, the simultaneous disruption of routine care and access to health care in acute and community settings for the non- COVID-19 population could potentially reduce the overall use of antimicrobials. The wider implications of the pandemic for health and social care systems must be characterized, and data must be gathered to inform and update national strategies that address the long-term challenges of AMR while preserving access to effective drugs. The emergence of SARS-CoV-2 has required an un- precedented response to control the spread of the infection and protect the most vulnerable within society. Whilst the pandemic has focused society on the threat of emerging infections and hand hygiene, certain infection control and antimicrobial stewardship policies may have to be relaxed. It is unclear whether the unintended consequences of these changes will have a net-positive or -negative impact on rates of antimicrobial resistance. Furthermore, the urgent focus must be on controlling this pandemic and sustained efforts to address the longer-term global threat of antimicrobial resistance should not be overlooked. Antimicrobials have several roles during the pandemic at the individual and organizational impact. Agents are being explored in clinical trials as potential direct therapies for SARS-CoV-2 such as azithromycin and antivirals such as lopinavir [10,11] and ritonavir [10,11], and remdesivir [12]. Antimicrobials are commonly prescribed for the management of presumptive or confirmed bacterial co-infection directly related to COVID-19, cooccurring at the time of infection, or associated with health care, such as prolonged admission to critical care. Combined with a lack of rapid diagnostic and decision support tools to support the clinical management of COVID-19, increased unnecessary antimicrobial use occurs [13-15]. Direct risks to the patient include side effects and adverse drug events. Increases in unnecessary antimicrobial use potentiates future risk of AMR through driving selection of multi-drug resistant (MDR) organisms and car-bapenemase producing Enterobacteriaceae [16,17]. Meanwhile, the impact of the pandemic on antimicrobial prescribing and AMR in the community remains largely unknown. Sustained public campaigns on the role of antibiotics in viral infections as well as the increased use of telemedicine may also reduce antimicrobial prescribing and AMR, but this is yet to be described [18]. In addition to the service changes, some routine infection control practices may have been compromised, including isolation of patients with MDR organisms as well as the redeployment of diagnostics to focus on SARS-CoV-2 in place of routine screening and surveillance for resistant bacteria, other respiratory viruses and healthcare-associated infections. However, emphasis on the role of hand hygiene may reduce transmission of AMR and diseases that would drive antibiotic use [3]. Moreover, the pressure on finite resources, such as medications (including antimicrobials) and high volume needs such as personal protective equipment for patients with COVID-19 may deplete availability of resources for the prevention and management of other infectious diseases. Following the initial global surge in COVID-19 cases, further surges are expected until control measures, such as vaccination, are introduced.
Methods
This study was performed following WHO guidelines using an online tool for evidence synthesis and was conducted to identify common bacterial/fungal co-infections reported in patients diagnosed with coronavirus infections since January 1st 2020. The MEDLINE and EMBASE databases were searched from 1st January 2020 to 2nd October 2020 using a combination of broad-based (and wildcard) search criteria including, coronavirus, COVID-19, SARS-1, MERS, bacterial, co-infection, antibiotics, AMR, MDR. Given the rapidly evolving nature of the literature on SARS-COV-2, journal advanced articles in leading infection journals and bibliographies of relevant articles were also reviewed. Covidence review gets out of the way so that we can bring the best evidence to the world, more quickly.
Results
Ongoing antimicrobial stewardship for co-infection and antimicrobial usage
We strongly believe co-infections play a significant role during COVID-19. Chronic obstructive pulmonary disease (COPD) is one of comorbidities associated with severe COVID-19. COPD patients are colonized by bacterial pathogens even at the stable phase of the disease, making it likely that SARS- CoV-2 infection occurs in patients already colonized with bacteria. Secondly, the very likely possibility exists that severe COVID-19 patients could be subsequently or co-incidentally infected by bacteria. The median hospital stage of COVID-19 patients is 7 days but can reach up to 14 days or even longer, and the risk of a hospital-associated pneumonia increases significantly the longer the hospitalization period. As shown in Figure 1, SARS-CoV-2 virulence factors interact with the lungs and evoke an immune response. Co-infection may result in an exuberant inflammatory response. It is also plausible that the type of immune response induced by SARS-CoV-2 may enable bacteria to flourish in the lungs. On the other hand, bacterial colonization may predispose to SARS-CoV-2 infection because the innate immune host defenses may be down-regulated enabling virus survival, growth and pathology. Co-infection may exacerbate the tissue damage; and the exuberant inflammatory response may further amplify the lung damage triggered by SARS-CoV-2. Addition to it, we demonstrate that NHC potently inhibits the divergent -CoVs MHV and MERS-CoV [19]. These data are consistent with a mode of action, as evidenced by a decrease in specific infectivity and an increase in G: A and C: U transition mutations present at low frequencies across the genome after treatment with NHC. We also demonstrate that robust resistance to NHC is difficult to achieve in both MHV and MERS-CoV as shown in Figure 2.
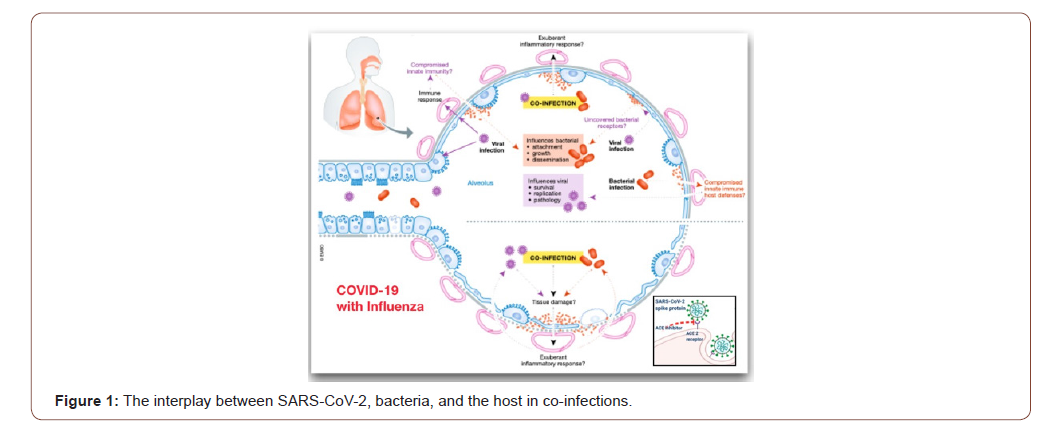
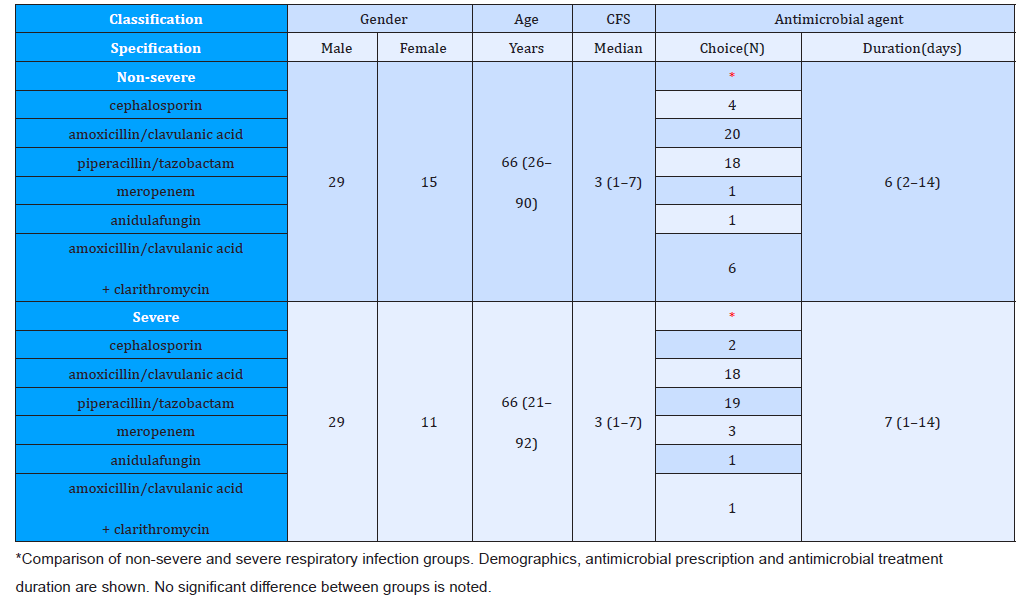
Consequently, bacterial respiratory co-infection in the setting of SARS-CoV-2 infection remains poorly described. A description of co-infection and antimicrobial usage is needed to guide ongoing antimicrobial stewardship [20-23]. The rate of bacterial coinfection in SARS-CoV-2 is low. De- spite this, prolonged courses of antimicrobial therapy were primary required in further cohort study. We recommend active antimicrobial stewardship in COVID-19 cases to ensure appropriate antimicrobial prescribing. This study reflects antimicrobial prescribing practices during the COVID-19 pandemic in the absence of an active AMS team. We demonstrate prolonged duration of antimicrobial treatment despite an alternative diagnosis and absence of evidence for bacterial co-infection as presented in Table 1. We suggest that AMS interventions are required in any ongoing COVID-19 infections, particularly focusing on discontinuing empirical therapy in the setting of negative cultures. Furthermore, the latest update of WHO’s interim guidance on the clinical management of COVID-19 incorporates antibiotic stewardship principles with specific recommendations [24]. The guidance further states that empiric antibiotic bacterial pneumonia treatment can be considered in older people residing in long-term care facilities. As these are nonhospitalized patients, antibiotics within WHO’s AWaRe (access, watch, reserve) classification of antibiotics categorized as access, such as co-amoxicillin, should preferably be administered.
Table 1: Distribution of ever-married females by age group and gravida and para, Kampung Peninjau Lama, December 2013.
Potential long-term impact of COVID-19 on antimicrobial use and AMR
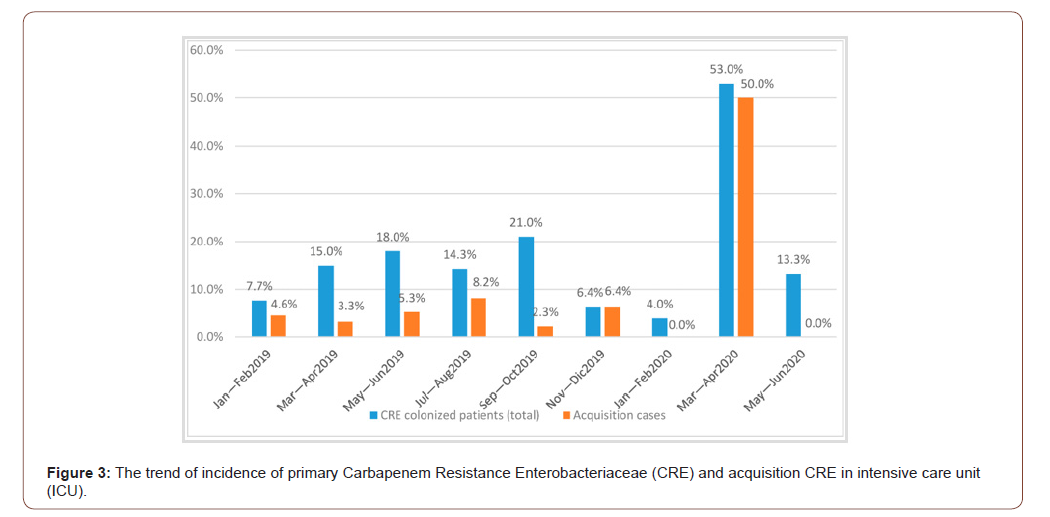
Consideration needs to be given to the impact of the COVID-19 pandemic and vaccination has been identified as a potential key intervention for reducing antimicrobial usage and therefore drugresistant infections. There is a risk that the pandemic will lead to major disruptions to immunization services, with major longterm consequences, including secondary outbreaks of vaccinepreventable diseases. The impact on control of MDR tuberculosis as a result of demands on health resources is likely to disproportionately impact low-income and middle-income countries, placing strain on fragile and overstretched health-care systems [25-27]. Particular attention was given to infection control practices with hand hygiene intervention, definition by checklist and continuous training on contact precautions for CRE carriers [28]. We also recommend active surveillance through systematic screening with rectal swab and clinical culture to search patients colonized by CRE, performed on all new ICU admissions and tested again on the seventh day of hospitalization (see Figure 3). The surveillance test to detect CRE on rectal swab was a culture-based method with a direct inoculation into specific selective chromogenic media. It was reported that growing colonies were identified by Vitek-MS, matrix-assisted laser desorption [29]/ionization time-of-flight (MALDI-TOF, bioMeriéux) [30], and tested for carbapenems resistance [31,32] and antibiotic susceptibility using the Vitek 2 system (bioMerieux, Marcy-L’Étoile, France) [33]. Enterobacteriaceae colonies [34] were also analyzed by immunochromatography test, for the identification of OXA-48- like, OXA carbapenemase (OXA-163) [35], Klebsiella Pneumoniae [36], Carbapenemase (KPC) [37], New Delhi Metallo-beta-lactamase (NDM) [38], and Verona Imipenemase (VIM) carbapenemases [39].
To mitigate the potential long-term impact of COVID-19 on antimicrobial use and AMR, health and social care systems must act to ensure that knowledge and evidence are effectively gathered to rapidly shape understanding and inform action. Evaluation of the impact of the pandemic on antimicrobial usage, AMR and access to effective antimicrobial treatments is essential to support the development of mitigation strategies. Clinical treatment guidelines to limit unnecessary antimicrobial exposure, measures to maintain routine surveillance of AMR and review of national policies that do not neglect essential public health programs in tuberculosis control and immunization delivery are essential. These strategies need to be supported by responsive national and international action plans that embed AMR as a priority in the post-COVID-19 era. At the national level, diagnostics and decision support should be developed to differentiate secondary bacterial infection during COVID-19. The role of specific biomarkers such as procalcitonin warrants further investigation [40]. Linkage of data to artificial intelligence-driven decision support software may provide added benefit. Antimicrobial stewardship should become an essential part of the individual clinical teams practice. Broader stewardship engagement will promote wider adoption of interventions designed to optimize treatment and reduce inappropriate antimicrobial use. Within primary care, data are urgently required to evaluate the impact of COVID-19 on antimicrobial prescribing, antibiotic access and clinical outcomes of infection. Novel approaches to stewardship and optimizing prescribing including telemedicine must be explored. Rapid progress has been made globally in basic sciences, epidemiology, diagnostics and clinical research in tackling COVID-19. There is also a societal opportunity to capitalize on the experience and learning from the pandemic to improve existing infection control and to sustain the raised awareness of the importance of vaccines and effective antimicrobial therapy. Coordination at the national level is required to gather evidence on the complex consequences of COVID-19 to inform necessary actions to reduce the potential longer-term impact on AMR and access to effective antimicrobials. For future study it will be investigated that the ex- acerbation of antimicrobial resistance (AMR), viral persistence and hyper-activation of the immune system by COVID-19 may be linked to inactive anti-aging genes. In the developing world plasma bacterial lipopolysaccharides (LPS) have increased and associated with repression of the anti-aging gene Sirtuin 1 the COVID-19 viral persistence and hyper-activation of the immune system is possibly extended with Sirtuin 1 gene inactivation. Activators of Sirtuin 1 may be critical to prevent the severity of the COVID-19 infection and exacerbation of AMR [41- 43].
Future design of synthetic vaccines as antagonists of COVID-19 infection
In the midst of the SARS-CoV-2 or COVID-19 outbreak the need for research into the development of antiviral agents is brought into sharp focus worldwide for scientists, governments and the public alike [44]. This special issue includes primary research into new antiviral agents with activity against a range of clinically and economically important viruses. SARS-CoV-2 is a newly emerged β-Coronavirus [45] with sustained human-to-human transmission via the respiratory tract. Influenza viruses are another group of respiratory viruses, which account for 300–600 K deaths annually and represent an ever-present pandemic threat. Emergence of drug-resistant influenza strains is driving research into new anti-influenza drug candidates with different modes of action additionally [46].
Computational Methods
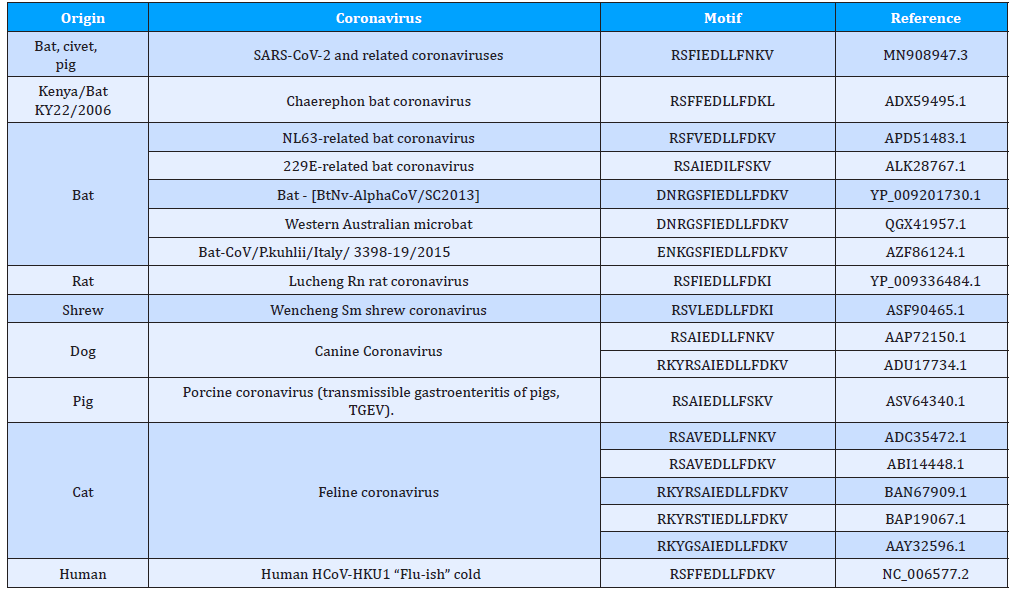
The main methods are essentially standard bioinformatics approaches as used and some methods, e.g. rules for epitope prediction, are best discussed in this study context [47-49]. The Q-UEL methods (see Figure 4) specifically for bioinformatics are discussed [50-54], and those for computational chemistry and docking of compounds are those using KRUNCH as described in Figure 5. They are somewhat unorthodox by focusing on heuristics to handle the multiple energy minimum problem, but the end effect is probably similar to that of long runs using high grade molecular dynamics calculations, given opportunities for calibration. Epitope predictions lie in more traditional one dimensional bioinformatics, and in this paper but briefly, it is in part because sections predicted with runs of tendency to be immunogenic even if they are incorrect as structure predictions. However, charged residues in α-helices and β-sheets are believed to be occasionally B-epitopes, and short sections extended chain can effectively imply loops. The core and initial rules for B-epitope prediction used in the present study [55,56]. A main conclusion is that peptide KRSFIEDLLFNKV remains of special interest as well conserved across coronaviruses. Some modifications of the RSFIEDLLFNKV motif in CoVids of mammalian hosts summarized in Table 2. Other sites and other proteins of the virus may, of course, emerge as the solutions to this formidable problem. All aspects of the virus must be considered. However, even the ACE2 binding domain is significantly more prone to accepted mutations (see Figure 5). The recurrence of the core features of the KRS- FIEDLLFNKV motif over so many diverse species reminds us of zoonotic origins, and it might be recalled that we consider that many and perhaps all plagues of mankind might ultimately be of ani- mal origin (see Table 2).
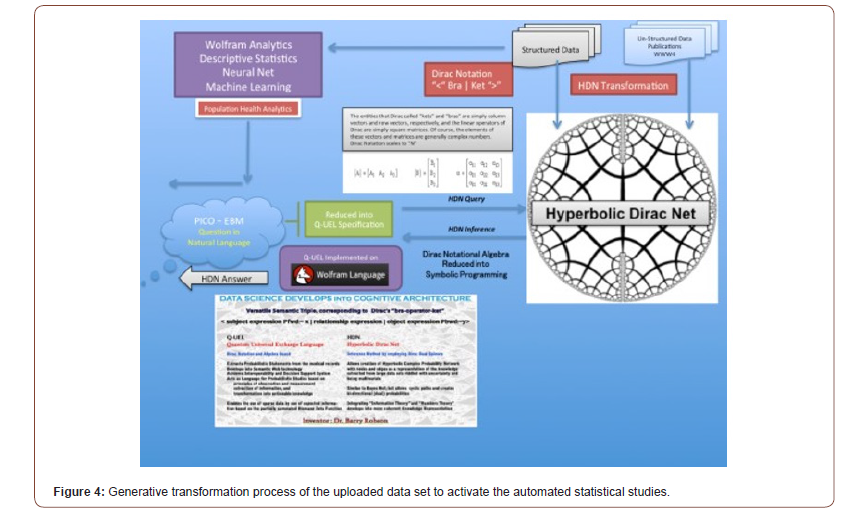
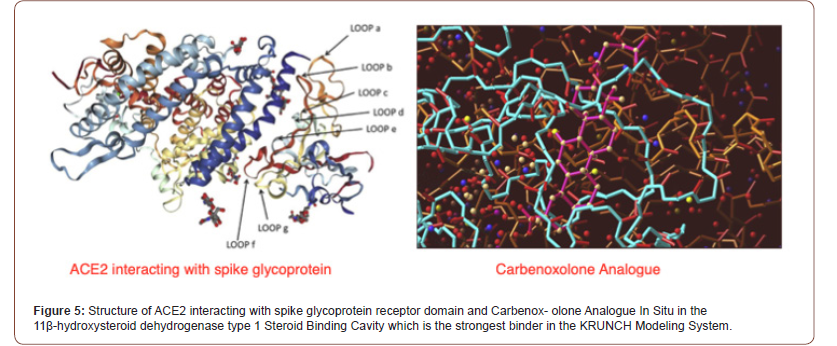
Table 2: Coronaviruses from mammalian hosts modified in RSFIEDLLFNKV motif of epitopes.
An algorithm-based method for overcoming resistance to antiviral agents
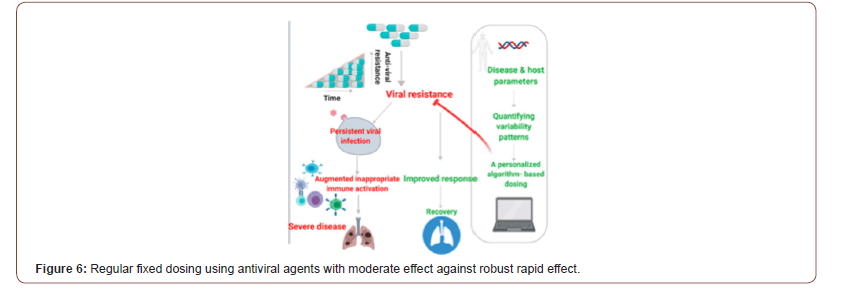
COVID-19 disease pathogenesis is characterized by an initial virus-mediated phase, followed by inappropriate hyper-activation of the immune system leading to organ damage. Targeting of the SARS-CoV-2 viral receptors is being explored as a therapeutic option for these patients. In previous paper, we summarize several potential receptors associated with the infectivity of SARS-CoV-2 and discuss their association with the immunemediated inflammatory response [57]. In addition to it, this paper is presented the potential for the development of resistance towards antiviral drugs. An algorithm-based platform to improve the efficacy of and overcome resistance to viral receptor blockers through the introduction of personalized variability is described in Figure 6. This method is designed to ensure sustained antiviral effectiveness when using SARS-CoV-2 receptor blockers [58]. The introduction of personalized variability to counter resistance to antiviral treatments. Regular, fixed dosing using an antiviral agent with robust and rapid effects is expected to lead to viral clearance, while fixed dosing regimens of moderately effective drugs may lead to drug resistance, viral persistence, an augmented hyper-activation of the immune system, and severe disease. The introduction of an algorithm-based dosing method based on quantified variability patterns derived from disease and host parameters improves the response to antiviral drugs.
Discussion
One dimension we feel has not received the necessary attention is the impact of COVID-19 on AMR. Globally, the COVID-19 pandemic is superimposed on the ongoing pandemic of multi-drug resistant bacteria. While we do not argue against this practice, we stress that it is important to note that some of them may content additional chemicals that do not add much in terms of protection but instead may fuel bacterial antimicrobial resistance. Bacteria exploit efflux pumps to develop resistance against disinfectants, and these same efflux pumps contribute to AMR. In any case, it is essential that the public adhere to proper use to avoid the selection of bacteria with increased tolerance/ resistance to antimicrobials. Thus, the development of antiviral agents is brought into sharp focus worldwide against MDR and AMR.
Conclusion
Many studies reporting on patients hospitalized with COVID-19 reveals widespread use of antimi- crobial therapies as part of the package of clinical care in some countries. Recent letter highlighted a need to prospectively monitor co-infections in patients with COVID-19 to understand whether co- infection affects disease progression, and to enable antimicrobial stewardship. It is of the utmost importance that the potential of the global pandemic to increase AMR is taken seriously. Therefore, new drug discovery for COVID-19 should be taken into consideration.
Transparency Declarations
None to declare.
Acknowledgment
We are grateful to Authorities in Irish Publishing on Academic Journal of Immunology and Micro- biology for complete wavier towards publication of my article at this issue.
References
- Beović B, Doušak M, Ferreira-Coimbra J, Nadrah K, et al. (2020) Antibiotic use in pa- tients with COVID-19: a 'snapshot' Infectious Diseases International Research Initiative (ID- IRI) The Journal of antimicrobial chemotherapy, dkaa, pp. 326.
- Ma C, Sacco MD, Hurst B, Townsend JA, et al. (2020) Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main pro- Cell research, 30(8): 678-692.
- Wilson LA, Rogers Van Katwyk S, Fafard P, Viens AM, Hoffman SJ (2020) Lessons learned from COVID-19 for the post-antibiotic future. Globalization and health 16(1): 94.
- Matsunaga N, Hayakawa K, Terada M, Ohtsu H, Asai Y, et al. (2020) Clinical epidemiology of hospitalized patients with COVID-19 in Japan: Report of the COVID-19 REGISTRY Clinical infectious diseases: an official publication of the Infectious Diseases Society of America, ciaa, pp. 1470.
- Kost GJ (2020) Geospatial Spread of Antimicrobial Resistance, Bacterial and Fungal Threats to COVID-19 Survival, and Point-of-Care Solutions. Archives of pathology & laboratory medicine, pp. 10.5858.
- Usman M, Farooq M, Hanna K (2020) Environmental side effects of the injudicious use of antimicrobials in the era of COVID-19. The Science of the total environment 745:
- Nieuwlaat R, Mbuagbaw L, Mertz D, Burrows L, Bowdish D, et al. (2020) COVID-19 and Antimicrobial Resistance: Parallel and Interacting Health Emergencies. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America, ciaa, pp. 773.
- Rawson TM, Moore L, Castro Sanchez E, Charani E, Davies F, et al. (2020) COVID-19 and the potential long-term impact on antimicrobial resistance. The Journal of antimicrobial chemotherapy 75(7): 1681-1684.
- Rawson TM, Ming D, Ahmad R, Moore L, Holmes AH (2020) Antimicrobial use, drug-resistant infections and COVID-19. Nature reviews. Microbiology 18(8): 409-410.
- Vecchio G, Zapico V, Catanzariti A, Carboni Bisso I, Las Heras M (2020) Efectos adversos de lopinavir/ritonavir en enfermedad grave por coronavirus (COVID-19) [Adverse effects of lopinavir/ritonavir in critically ill patients with COVID-19]. Medicina 80(5): 439-441.
- Alvarez JC, Moine P, Davido B, Etting I, Annane D, et al. (2020) Population pharma- cokinetics of lopinavir/ritonavir in Covid-19 patients. European journal of clinical pharmacology, pp. 10.1007.
- Shih RD, Maki DG, Hennekens CH (2020) Remdesivir for coronavirus 2019 (COVID-19): More promising but still unproven. Contemporary clinical trials communications, pp. 100663.
- Ali E, Ziglam H, Kohla S, Ahmed M, Yassin M (2020) A Case of Fulminant Liver Failure in a 24-Year-Old Man with Coinfection with Hepatitis B Virus and SARS-CoV-2. The American journal of case reports 21: e925932.
- de Koff EM, van Houten MA, Sanders E, Bogaert D (2020) Severity of Respiratory Infections with Seasonal Coronavirus Is Associated With Viral and Bacterial Coinfections. The Pediatric infectious disease journal.
- Song W, Jia X, Zhang X, Ling Y, Yi Z (2020) Co-infection in COVID-19, a cohort The Journal of infection S0163-4453(20): 30648-30654.
- Matouk AE (2020) Complex dynamics in susceptible-infected models for COVID-19 with multi-drug resistance. Chaos, solitons, and fractals 140: 110257.
- Bhuiyan FR, Howlader S, Raihan T, Hasan M (2020) Plants Metabolites: Possibility of Natural Therapeutics against the COVID-19 Pandemic. Frontiers in medicine 7: 444.
- Yoon EJ, Tong D, Anton GM, Jasinski JM, Claus CF, et al. (2020) Patient Satisfaction with Neurosurgery Telemedicine Visits During the COVID-19 Pandemic: A Prospective Cohort World neurosurgery S1878-8750(20): 32189-32196.
- Agostini ML, Pruijssers AJ, Chappell JD, Gribble J, Xiaotao Lu, et al. (2019) Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. Journal of virology 93(24): e01348-19.
- Lynch C, Mahida N, Gray J (2020) Antimicrobial stewardship: a COVID casualty? The Journal of hospital infection S0195-6701(20): 30462-X.
- Riad A, Gad A, Hockova B, Klugar M (2020) Oral Candidiasis in Non-Severe COVID-19 Patients: Call for Antibiotic Stewardship. Oral surgery.
- Stevenson DR, Sahemey M, Cevallos Morales J, Martín-Lázaro J, Buchanan R, et al. (2020) Improving Antimicrobial Stewardship in Critically-Ill Patients with COVID-19. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America, ciaa, pp. 1559.
- van Berkel M, Kox M, Frenzel T, Pickkers P, Schouten J (2020) Biomarkers for antimicrobial stewardship: a reappraisal in COVID-19 times? Critical care (London, England) 24(1): 600.
- Torres I, Artaza O, Profeta B, Alonso C, Kang J (2020) COVID-19 vaccination: returning to WHO's Health For All. The Lancet. Global health 8(11): e1355-e1356.
- Vilbrun SC, Mathurin L, Pape JW, Fitzgerald D, Walsh KF (2020) Case Report: Multidrug-Resistant Tuberculosis and COVID-19 Coinfection in Port-au-Prince, Haiti. The American journal of tropical medicine and hygiene.
- Nindrea RD, Sari NP, Harahap WA, Haryono SJ, Kusnanto H, et al. (2020) Survey data of multidrug-resistant tuberculosis, Tuberculosis patients characteristics and stress resilience during COVID-19 pandemic in West Sumatera Province, Indonesia. Data in brief 32: 106293.
- Alagna R, Besozzi G, Codecasa LR, Gori A, Migliori GB, et al. (2020) Celebrating World Tuberculosis Day at the time of COVID-19. The European respiratory journal 55(4): 2000650.
- Yan L, Sun J, Xu X, Huang S (2020) Epidemiology and risk factors of rectal colonization of carbapenemase producing Enterobacteriaceae among high-risk patients from ICU and HSCT wards in a university hospital. Anti-microbial resistance and infection control 9(1): 155.
- Akgül Ö, Söyletir G, Ülger Toprak N (2020) Antimicrobial Susceptibility of Pathogenic Gram-positive Anaerobic Cocci: Data of a University Hospital in Turkey. Mikrobiyoloji bulteni 54(3): 404-417.
- Matsuo T, Mori N, Kawai F, Sakurai A, Toyoda M, et al. (2020) Vagococcus fluvialis as a causative pathogen of bloodstream and decubitus ulcer infection: Case report and systematic review of the literature. Journal of infection and chemotherapy: official journal of the Japan Society of Chemotherapy S1341-321X(20): 30339-30341.
- Porretta AD, Baggiani A, Arzilli G, Casigliani V, Mariotti T, et al. (2020) Increased Risk of Acquisition of New Delhi Metallo-Beta-Lactamase-Producing Carbapenem-Resistant Enterobacterales (NDM-CRE) among a Cohort of COVID-19 Patients in a Teaching Hospital in Tuscany, Pathogens (Basel, Switzerland) 9(8): 635.
- Lima WG, Brito J, da Cruz Nizer WS (2020) Ventilator-associated pneumonia (VAP) caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19: Two problems, one solution? Medical hypotheses 144: 110139.
- Aprile A, Scalia G, Stefani S, Mezzatesta ML (2020) In vitro fosfomycin study on concordance of susceptibility testing methods against ESBL and carbapenem resistant Enter Journal of global antimicrobial resistance S2213-7165(20): 30254-X.
- Xiaomin S, Yiming L, Yuying Y, Zhangqi S, Yongning W, et al. (2020) Global impact of mcr-1-positive Enterobacteriaceae bacteria on "one health". Critical reviews in microbiology ,p. 1-13.
- Gelmez GA, Can B, Hasdemir U, Soyletir G (2020) Evaluation of two commercial methods for rapid detection of the carbapenemase producing Klebsiella pneumoniae. Journal of microbiological methods 178: 106084.
- Gong L, Tang N, Chen D, Sun K, Lan R, et al. (2020) A Nosocomial Respiratory Infection Outbreak of Carbapenem Resistant Escherichia coli ST131 With Multiple Transmissible bla KPC-2 Carrying Plasmids. Frontiers in microbiology 11:
- Chen Q, Lin Y, Li Z, Lu L, Li P, et al. (2020) Characterization of a New Transposon, Tn6696, on a blaNDM-1-Carrying Plasmid From Multidrug Resistant Enterobacter cloacae ssp. dissolvens in China. Frontiers in microbiology 11:
- Gill CM, Lasko MJ, Asempa TE, Nicolau DP (2020) Evaluation of the EDTA Modified Carbapenem Inactivation Method for Detecting Metallo-β-Lactamase-Producing Pseudomonas aeruginosa. Journal of clinical microbiology 58(6): e02015-19.
- Mohamed S, Abdelhaffez A, Abd El-Aziz N (2020) Serum Procalcitonin in Patients With Combined Lung Cancer and Idiopathic Pulmonary Fibrosis (LC-IPF). Cureus 12(8): e9507.
- Gonçalves BC, Lopes Barbosa MG, Silva Olak AP, Faccin-Galhardi LC (2020) Antiviral therapies: advances and perspectives. Fundamental & clinical pharmacology.
- Goli M (2020) Review of novel human β-coronavirus (2019-nCoV or SARS-CoV-2) from the food industry perspective-Appropriate approaches to food production technology. Foodscience & nutrition.
- Ginex T, Luque FJ (2020) Searching for effective antiviral small molecules against influenza A virus: A patent review. Expert opinion on therapeutic patents.
- Oladipo EK, Ajayi AF, Ariyo OE, Onile SO, et al. (2020) Exploring Surface Glycoprotein to Design Multi-Epitope Vaccine Against Covid-19. Informatics in medicine unlocked 21: 100438.
- Dawood AA (2020) Glicosilación, sitios de unión de ligandos y variaciones antigénicas entre la glicoproteína de membrana del COVID-19 y los coronavirus asociados [Glycosylation, Ligand Binding Sites and Antigenic Variations Between Membrane Glycoprotein of COVID-19 and Related Coronaviruses]. Vacunas.
- Dhall A, Patiyal S, Sharma N, Usmani SS, Raghava G (2020) Computer aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19. Briefings in bioinformatics, bbaa, pp. 259.
- Robson B (2020) Extension of the Quantum Universal Exchange Language to precision medicine and drug lead discovery. Preliminary example studies using the mitochondrial genome. Computers in biology and medicine 117:
- Robson B (2016) Studies in using a universal exchange and inference language for evidence based medicine. Semi-automated learning and reasoning for PICO methodology, systematic review, and environmental epidemiology. Computers in biology and medicine 79: 299-323.
- Robson B, Boray S (2016) Data-mining to build a knowledge representation store for clinical decision support. Studies on curation and validation based on machine performance in multiple choice medical licensing examinations. Computers in biology and medicine 73: 71-93.
- Robson B, Boray S (2015) Implementation of a web based universal exchange and inference language for medicine: Sparse data, probabilities and inference in data mining of clinical data repositories. Computers in biology and medicine 66: 82-102.
- Robson B (2015) POPPER, a simple programming language for probabilistic semantic infer- ence in medicine. Computers in biology and medicine 56: 107-123.
- Singh A, Thakur M, Sharma LK, Chandra K (2020) Designing a multiepitope peptide based vaccine against SARS-CoV-2. Scientific reports 10(1): 16219.
- Romero López JP, Carnalla Cortés M, Pacheco Olvera DL, Jiménez Zamudio L (2020) A bioinformatic prediction of antigen presentation from SARS-CoV-2 spike protein revealed a theoretical correlation of HLA-DRB1*01 with COVID-19 fatality in Mexican population: An ecological approach. Journal of medical virology.
- Gelman R, Bayatra A, Kessler A, Schwartz A, Ilan Y (2020) Targeting SARS-CoV-2 receptors as a means for reducing infectivity and improving antiviral and immune response: an algorithm-based method for overcoming resistance to antiviral agents. Emerging microbes & infections 9(1): 1397-1406.
- Sadria M, Layton AT (2020) Use of Angiotensin Converting Enzyme Inhibitors and Angiotensin II Receptor Blockers During the COVID-19 Pandemic: A Modeling Analysis. PLoS computational biology 16(10): e1008235.
-
Hyunjo Kim, Jae-Hoon Song. An Algorithm-Based Computational Method for Overcoming Worldwide Antimicrobial Resistance Exacerbation in COVID-19 Pandemic Era. Acad J Microbiol & Immun. 1(1): 2020. AJMI.MS.ID.000505.
-
Antimicrobials; COVID-19; Chronic obstructive pulmonary disease; BioMeriéux; Lipopolysaccharides
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






