 Mini Review
Mini Review
Salivary Biomarkers in Gastric Cancer: A Non-Invasive Biomarkers for Early Detection and Diagnostics
Anju R Nath and Jeyakumar Natarajan*
Department of Bioinformatics, Bharathiar University, India
Jeyakumar Natarajan, Data Mining and Text Mining Laboratory, Department of Bioinformatics, Bharathiar University, Coimbatore 641046, India.
Received Date: June 26, 2020; Published Date: July 08, 2020
Abstract
Gastric cancer is one of the aggressive tumors which has a poor prognosis due to the late detection. It is asymptomatic cancer and most of the cases gastric cancer are diagnosed at advanced stages. From the usual invasive methods of screening, the use of saliva as a non-invasive diagnostic method is gaining much attention. Nowadays researches have shed light on the use of saliva as a diagnostic tool and many studies have identified various salivary biomarkers for the early detection of gastric cancer. But the lack of validated biomarkers in this field can be considered as a drawback, which can be dealt with the enhancement of preclinically validated biomarkers by assimilating them to credible biomarker panels.
Introduction
Gastric cancer (GC) is one of the most common malignancies and the third leading cause of cancer-related death worldwide [1]. According to the International Agency for Research on Cancer, in the year 2018 alone, 1,33,701 cases have been reported all over the world, i.e. 6.1% of the total number of cases. Alcohol intake, smoking and dietary factors such as high salt intake and genetics, are suggested to be associated with the development of gastric cancer [2]. GC is an aggressive tumor which carries a poor prognosis due to advanced- stage detections. Thus, it is critical to developing a method that can diagnose the disease at an early stage to allow for better treatment options. Currently, for GC, there are limited early diagnostic and screening methods such as biopsy, endoscopy, enhanced CT and upper gastrointestinal angiography [3]. To identify a non-invasive means of early diagnosis or screening for gastric cancer is important in the current scenario. As a noninvasive diagnostic technique, diagnostics with saliva has become an engaging indicator to assess the physiological and pathological states [4-7]. Few studies have investigated the relevancy of salivary biomarkers in the detection of gastric cancer. The various categories of salivary biomarkers such as small protein biomarkers, small metabolite biomarkers exRNA biomarkers, D-amino acid biomarkers etc. were discussed in this mini-review.
Discussion
Table 1: Salivary diagnostic biomarker candidates for gastric cancer.
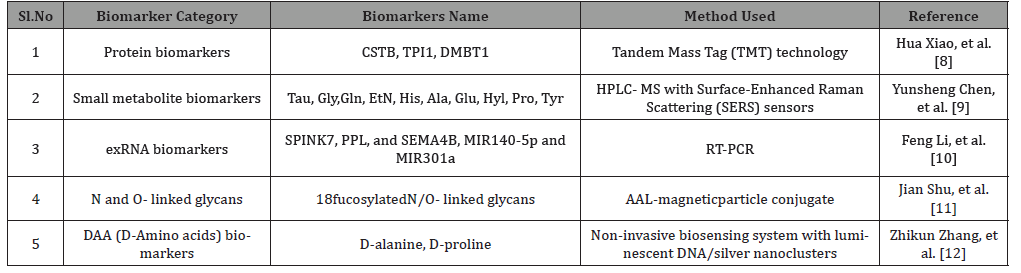
The salivary biomarkers are broadly classified into four categories such as protein biomarkers, small metabolite biomarkers, exRNA biomarkers, and D-amino acid biomarkers. Each category and the biomarkers related to the category are illustrated in Table 1 with the methods used. This is followed by a brief discussion of each biomarker (Table 1).
Protein Biomarkers: Amoung the protein biomarkers, CSTB (Cystatin B promotes cell proliferation and migration and also suppresses apoptosis, thus, encouraging the GC development [13]. TPI1 (triosephosphate isomerase) acts as an anti-drug resistance agent in GC [14], and DMBT1 (deleted in malignant brain tumors 1 protein) upregulation can be found in across all types of GCs. Tandem Mass Tag (TMT) technology was used to analyse and compare salivary proteins. Selected proteins were verified using ELISA [8].
Small metabolite biomarkers: From the saliva ten amino acids (Tau, Gly,Gln, EtN, His, Ala, Glu, Hyl, Pro, Tyr) were identified as small metabolite biomarkers using high-performance liquid chromatography-mass spectrometry (HPLC- MS) and the ten amino acids were detected in saliva using surface-enhanced Raman Scattering (SERS) sensors. Rather than individual amino acids, the combination of these biomarkers helps in distinguishing early GC patients from advanced GC patients [9].
exRNA biomarkers: All three SPINK7, PPL, and SEMA4B were found downregulated in GC. These genes are expressed mainly in Esophageal squamous carcinoma, lung cancer etc. The MIR140- 5p can suppress the proliferation and invasion of GC by regulating YES1[15]. Also, upregulation of MIR301a can promote cell proliferation and invasion in GC by targeting RUNX3 [16]. From the transcriptome and miRNA profiles of saliva samples, the exRNAs were verified using RT-qPCR [10].
N and O-linked glycans: A total of 18 fucosylated N/O-linked glycans were found in saliva samples of GC patients. Alterations in glycans affect the glycoproteins which may contribute to the malignant transformation in GCThefucosylated glycans were isolated from the samples using the AAL- magnetic particle conjugates [11].
DAA (D-amino acid) biomarkers: Due to the higher concentration of D-alanine and D- proline in GC they were selected as biomarkers for GC diagnosis. Then using a non-invasive biosensing system with luminescent DNA/silver nanoclusters these DAAs were identified from the saliva sample of GC patients at early stage indicating the suitability of the method for early diagnosis of GC.
Conclusion
The cancer screening techniques intend to find cancers at an early stage. This includes blood test, DNA tests, scientific imaging etc. But most of them are invasive methods. Here, non-invasive cancer screening may be considered as a boon. The role of saliva as a non-invasive screening method in cancers is essential. Saliva is a body fluid which is a mixture of secretions from numerous salivary glands. The biological functions of saliva maintain oral and systemic health. Researchers are developing salivary diagnostic tools for the detection of oral and systemic diseases such as gastric cancer, colorectal cancer, Esophageal cancer etc. The salivary biomarkers are important non-invasive diagnostic biomarkers. But the FDA approved established salivary biomarkers may be stated to be absent. Researches are going on and many biomarkers have been diagnosed and tested preclinically. The amalgamation of the recognized biomarkers, i.e. a biomarker panel, might be a tremendous way to facilitate saliva as a biomarker for the early identification of cancers which include gastric cancers.
Acknowledgement
None.
Conflict of Interest
No conflict of interest.
References
- Ferlay J, Soerjomataram I, Dikshit R, Sultan E, Colin M, et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5): E359–E386.
- Kelley JR, Duggan JM (2003) Gastric cancer epidemiology and risk factors. J Clin Epidemiol 56:1-9.
- den Hoed CM, Kuipers EJ (2016) Gastric cancer: How can we reduce the incidence of this disease? Curr Gastroenterol Rep 18: 34.
- Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, et al. (2013) Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev 26: 781-791.
- Elashoff D, Hui Z, Jean R, Jianghua W, Hua X, et al. (2012) Prevalidation of Salivary Biomarkers for Oral Cancer Detection. Cancer Epidemiology, Biomarkers & Prevention 21(4): 664–672.
- Zhang L, James JF, Hui Z, David E, David A, et al. (2010) Salivary Transcriptomic Biomarkers for Detection of Resectable Pancreatic Cancer. Gastroenterology 138(3): 949–957.
- Zhang L, Hua X, Hui Z, Silverio S, Jay ML, et al. (2012) Development of Transcriptomic Biomarker Signature in Human Saliva to Detect Lung Cancer. Cellular and Molecular Life Sciences 69(19): 3341–3350.
- Xiao H, Yan Z, Yong K, Sung K, Jae JK, et al. (2016) Differential Proteomic Analysis of Human Saliva Using Tandem Mass Tags Quantification for Gastric Cancer Detection. Scientific Reports 6(1): 22165–22165.
- Chen Y, Shangli C, Aming Z, Jie S, Jie C, et al. (2018) Salivary Analysis Based on Surface-Enhanced Raman Scattering Sensors Distinguishes Early and Advanced Gastric Cancer Patients from Healthy Persons. Journal of Biomedical Nanotechnology 14(10): 1773–1784.
- Li F, Janice M, Kyoung-Mee K, Julie K, Tristan RG, et al. (2018) Discovery and Validation of Salivary Extracellular RNA Biomarkers for Noninvasive Detection of Gastric Cancer. Clinical Chemistry 64(10): 1513–1521.
- Shu J, Hanjie Y, Haoqi D, Jiaxu Z, Kun Z, et al. (2018) Identification of N- and O-linked Glycans Recognized by AAL in Saliva of Patients with Atrophic Gastritis and Gastric Cancer. Cancer Biomarkers 22(4): 669–681.
- Zhang Z, Yang L, Peifeng L, Lu Y, Xingyu J, et al. (2017) Non-Invasive Detection of Gastric Cancer Relevant D-Amino Acids with Luminescent DNA/Silver Nanoclusters. Nanoscale 9(48): 19367–19373.
- Zhang J, Zhen FS, Jin XH, Xiao GZ (2016) CSTB Downregulation Promotes Cell Proliferation and Migration and Suppresses Apoptosis in Gastric Cancer SGC-7901 Cell Line. Oncology Research 24(6): 487–494.
- Wang X, Yuanyuan L, Jinghua Y, Yongquan S, Mei L, et al. (2008) Identification of Triosephosphate Isomerase as an Anti-Drug Resistance Agent in Human Gastric Cancer Cells Using Functional Proteomic Analysis. Journal of Cancer Research and Clinical Oncology 134(9): 995–1003.
- Fang Z, Shuai Y, Ruochuan S, Shangxin Z, Min F, et al. (2017) MiR-140-5p Suppresses the Proliferation, Migration and Invasion of Gastric Cancer by Regulating YES1. Molecular Cancer 16(1): 139.
- Wang M, Chenglong L, Beiqin Y, Liping S, Jianfang L, et al. (2013) Overexpressed MiR-301a Promotes Cell Proliferation and Invasion by ‘Targeting RUNX3 in Gastric Cancer. Journal of Gastroenterology 48(9): 1023–1033.
-
Anju R Nath, Jeyakumar Natarajan. Salivary Biomarkers in Gastric Cancer: A Non-Invasive Biomarkers for Early Detection and Diagnostics. 2(5): 2020. ACRCI.MS.ID.000548.
-
Cancer, Salivary biomarkers, Tumors, Biopsy, Endoscopy, Concentration, Amino acids, Gastric cancer, Chromatography
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






