 Short Communication
Short Communication
Recurrent COQ8B-NUMBL Fusions are the Result of Transcriptional Readthrough Events Frequently Found in Solid Tumors
Diantha Terlouw1, Anne I M Vogel1, Ronald van Eijk1, Anne-Marie Cleton-Jansen1, Vincent T H B M Smit1, Hans Morreau1, Elisabeth M P Steeghs1, Dina Ruano1, Tom van Wezel1, 2*
1Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands
2Department of Pathology, The Netherlands Cancer Institute, Antoni van Leeuwenhoek Hospital, the Netherlands
Tom van Wezel, Department of Pathology, Leiden University Medical Center, Leiden, P1-36, Albinusdreef 2, 2333 ZA, The Netherlands
Received Date:March 29, 2024; Published Date:April 26, 2024
Abstract
Gene fusions are vital in oncogenesis, although they are rare and represent the sole drivers in a minor fraction of cancers. These fusions often stem from genomic rearrangements, notably translocations, which either enhance oncogenic functions or diminish tumor suppressor gene expression. Their detection and interpretation is important for diagnostic and therapeutic stratification efforts. In this context, the proposed ADCK4(COQ8B)- NUMBL gene fusion has been suggested as a frequent event driving cutaneous squamous cell carcinoma (cSCC) development. However, our analysis contests this assertion, demonstrating that COQ8B-NUMBL fusions observed in cSCC are not the result of genomic rearrangements but rather arise from transcriptional readthrough events between adjacent genes on chromosome 19. Our study, consisting of RNA sequencing data from a diverse range of solid tumors, challenges the (clinical) relevance of COQ8B-NUMBL fusions. Notably, we observe a substantial prevalence (40.8%) of these fusion transcripts across all tested tumor types. Furthermore, our findings reveal a significant co-occurrence (36.8%) between established oncogenic drivers and the COQ8B-NUMBL transcriptional readthrough event, suggesting this is not a driver in carcinogenesis. These insights highlight the importance of discerning between genuine genomic rearrangements and transcriptional artifacts to refine our understanding of oncogenic mechanisms. Overall, our study underscores the need for cautious interpretation of fusion events and advocates for comprehensive fusion analyses to elucidate their true biological significance in cancer pathogenesis.
Keywords: COQ8B-NUMBL; ADCK4; Gene Fusion; Transcriptional Readthrough; Oncogenesis; Solid Tumor
Abbreviations: cSCC: cutaneous squamous cell carcinoma
Introduction
Gene fusions play a critical role in oncogenesis, albeit that in only 1% of cancers, fusions are the sole driver of tumor development [1]. The most common driver fusions stem from a genomic rearrangement or translocation [2-3] and enhance oncogenic function or diminish tumor suppressor gene expression [1]. The detection and interpretation of oncogenic fusions is important for diagnostic and therapeutic stratification. For cutaneous squamous cell carcinomas (cSCC), the gene fusion ADCK4-NUMBL was suggested as a frequent event and driving tumorigenesis [4]. The official symbol for ADCK4 is COQ8B [5] and will be referred to as such. Egashira et al. [4] state that the fusion between COQ8B exon 14, and NUMBL exon 2 is the product of genomic rearrangement of chromosome 19. In this short communication, we contest this assertion and show that COQ8B-NUMBL fusions are instead the result of a transcriptional readthrough event and frequently found in solid cancer tissues, hence unlikely to be clinically relevant or even specific for cSSC.
Discussion
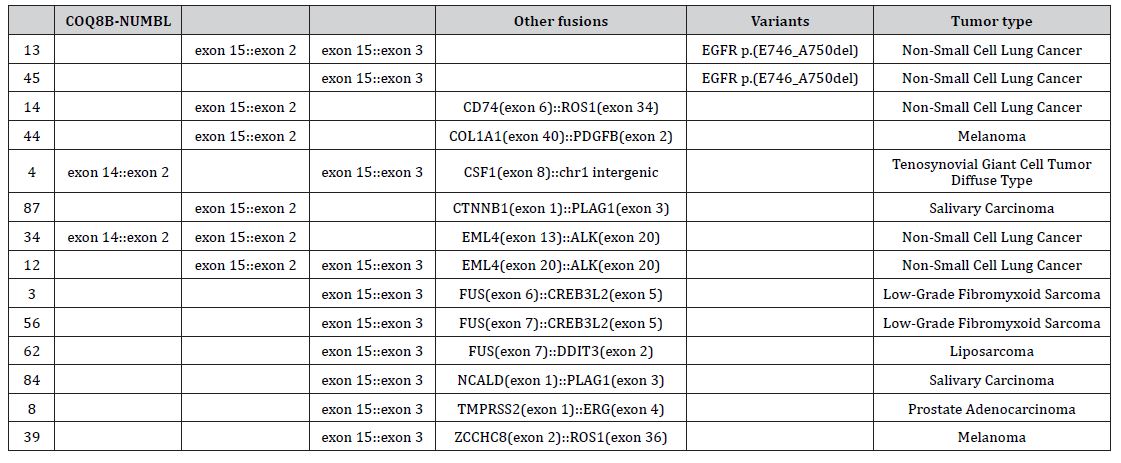
Based on the location within the human genome, the observed COQ8B-NUMBL fusion is not the result of a genomic rearrangement. COQ8B and NUMBL are adjacent genes, located on the complement strand at 19q13.2. As the distance between the two transcripts is only 879 nucleotides (Figure 1A), a transcriptional readthrough is the most likely explanation for the observed transcript, rather than an actual fusion of the two genes. During a transcriptional readthrough, transcription continues into the downstream gene instead of stopping at the canonical termination site, which can also result in alternative splicing variants. Contrarily, readthrough transcripts are observed in a wide range of cases [6] and are associated with cellular stress responses due to a decrease of transcriptional efficiency [6,7]. In order to further support our hypothesis in vivo, we investigated historical data from our routine molecular diagnostics. We recently performed RNA sequencing with the FusionPlex PanSolidTumor v2 18091 v1.0 panel covering NUMBL [8]. This cohort consisted of various tumors including non- small cell lung carcinomas (n=40), bone and soft tissue tumors (n=18), melanomas (n=9) thyroid tumors (n=4), salivary carcinomas (n=8), diffuse gliomas (n=3), nerve sheath tumors (n=2), hepatic tumor (n=2), metastasis of primary tumors unknown (n=2), prostate carcinoma (n=1), renal carcinoma (n=1), endometrial carcinoma (n=1), myeloid neoplasm (n=1) and a glomus tumor (n=1). Our data shows that COQ8B-NUMBL fusion transcripts (Figure 1B) are not unique for cSCCs and can be found in a variety of solid tumors, such as non-small cell lung carcinomas, melanomas, and salivary carcinomas. In 93 cases recently sequenced, 38 (40.8%) of the samples presented with COQ8BNUMBL fusion transcripts. Interestingly, this frequency is close to the frequency of COQ8B-NUMBL fusions found by Egashira et al. (i.e. 35.3%) [4]. Since Egashira et al. used a different NGS strategy, it is likely that this frequency is indicative of the same readthrough artifacts and not an artifact specific to our panel. Fourteen (15.1%) of the analyzed tumor tissues contain multiple transcript fusions of COQ8B-NUMBL, indicative for a variety of splice variants of the readthrough transcript. While we detect transcripts involving exon 14 of COQ8B and exon 2 of NUMBL as reported by Egashira et al., in 92.1% of cases other fusion transcripts were detected. In fact, the most commonly found fusion transcript (71.1%) is the COQ8B exon 15:: NUMBL exon 3 fusion transcript.
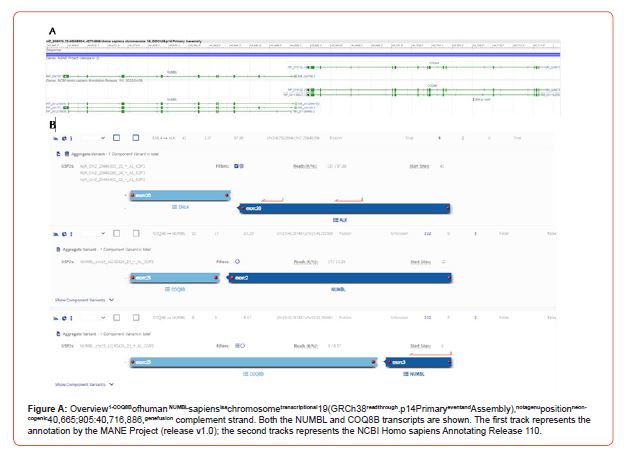
Importantly, our findings reveal a significant co-occurrence (36.8) between COQ8B- NUMBL fusions and established oncogenic drivers, both actual fusions and variants (Table 1), like CRTC1- MAML2 fusions in salivary carcinoma (specifically mucoepidermoid carcinomas), EML4-ALK fusions in non-small cell lung carcinomas and NAB2-STAT6 fusions in solitary fibrous tumors. This finding supports our hypothesis that COQ8B-NUMBL fusion transcripts do not act as drivers in carcinogenesis.
Table 1: analyses–TumorwithsamplestheFusionPlexwithCOQ8BPanSolid Tumor NUMBLtranscriptsv218091andv1.0estabpanelished [8] werefusicollectednsand betweenvariantsNovember 28, 2022 and January 9, 2023. Each sample was analyzed with Archer Analysis software 6.2.3 and the number of COQ8B- NUMBL fusions, other fusions and variants was recorded. Low confidence fusions and known artifacts were excluded from analysis. Tumor type was taken from the associated patient history and classified with the help of Oncotree [10].
Conclusion
In conclusion, we show that the COQ8B-NUMBL fusions do not result from genomic rearrangements, but from a transcriptional readthrough event of two adjacent genes. This event occurs in 40.8% (38/93) recently analyzed solid tumors, independent of tumor type, and is regularly found (36.8%) in combination with known oncogenic drivers. These insights highlight the importance of discerning between genomic rearrangements and transcriptional artifacts to refine our understanding of oncogenic mechanisms. (2022-04-08) and includes different transcript variants. Figure was generated by NCBI Sequence Viewer 3.47.0 [9]. B. Archer output of RNA sequencing of sample 12, two COQ8B-NUMBL transcripts cooccurred with the established EML4-ALK fusion in a non-small cell lung carcinoma.
Acknowledgements
The authors thank the Molecular Diagnostics unit of the pathology department for the technical support.
Conflict of Interest
The authors state no conflict of interest.
References
- Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, et al. (2018) Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep 23(1): 227-38 e3.
- Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, et al. (2018) TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 46(D1): D1144-D9.
- Luo JH, Liu S, Zuo ZH, Chen R, Tseng GC, Yu YP (2015) Discovery and Classification of Fusion Transcripts in Prostate Cancer and Normal Prostate Tissue. Am J Pathol 185(7): 1834-1845.
- Egashira S, Jinnin M, Makino K, Ajino M, Shimozono N, Okamoto S, et al. (2019) Recurrent Fusion Gene ADCK4-NUMBL in Cutaneous Squamous Cell Carcinoma Mediates Cell Proliferation. J Invest Dermatol 139(4): 954-957.
- Bethesda (MD): National Library of Medicine (US) NCfBI. COQ8B coenzyme Q8B [ Homo sapiens (human) ],
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA (2015) Widespread Inducible Transcription Downstream of Human Genes. Mol Cell 59(3): 449-461.
- Vilborg A, Steitz JA (2017) Readthrough transcription: How are DoGs made and what do they do? RNA Biol 14(5): 632-636.
- PALGA S Moleculaire bepaling.
- Rangwala SH, Kuznetsov A, Ananiev V, Asztalos A, Borodin E, Evgeniev V, et al. (2021) Accessing NCBI data using the NCBI Sequence Viewer and Genome Data Viewer (GDV). Genome Res 31(1): 159-169.
- Kundra R, Zhang H, Sheridan R, Sirintrapun SJ, Wang A, Ochoa A, et al. (2021) Onco Tree: A Cancer Classification System for Precision Oncology. JCO Clinical Cancer Informatics 2021(5): 221-30.
-
Diantha Terlouw, Anne I M Vogel1, Ronald van Eijk, Anne-Marie Cleton-Jansen, Vincent T H B M Smit, Hans Morreau, Elisabeth M P Steeghs, Dina Ruano, Tom van Wezel*. Recurrent COQ8B-NUMBL Fusions are the Result of Transcriptional Readthrough Events Frequently Found in Solid Tumors. Adv Can Res & Clinical Imag. 4(2): 2024. ACRCI.MS.ID.000585.
-
COQ8B-NUMBL; ADCK4; Gene Fusion; Transcriptional Readthrough; Oncogenesis; Solid Tumor
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






