 Research Article
Research Article
Looking at the TMEs from the Spatial Transcriptomics Perspective. What can we see?
Karla Paniagua1, Yufang Jin1 and Mario Flores1,2*
1Department of Electrical and Computer Engineering, University of Texas at San Antonio, USA
2Department of Biomedical Engineering, University of Texas at San Antonio, USA
Mario Flores, Department of Electrical Engineering, University of Texas at San Antonio, San Antonio, One UTSA Circle, TX 78249, USA.
Received Date: January 31, 2023; Published Date: February 21, 2023
Abstract
Recent advances in spatial transcriptomics technologies allow us to computationally visualize the tumor microenvironments (TMEs) with a never seen before resolution. These new platforms together with the use of artificial intelligence algorithms can pave the way for reformulation of questions we never even knew could be ask. In order to obtain some perspective of the use of these technologies, we reprocessed a public dataset of high-resolution spatial transcriptomics of non-small cell lung cancer (NSCLC) and perform computational visualization of the TMEs. Our results show two directions of research questions that can be ask using these promising technologies.
Keywords: Spatial; Transcriptomics; Tumor; Microenvironment; Computational; Visualization
Abbreviations: TMEs: Tumor microenvironments; DL: Deep Learning; spRNAseq: high-resolution single-cell spatially resolved transcriptomics; SMI: Spatial Molecular Imager; FFPE: formalin-fixed paraffin-embedded; FF: fresh frozen; DGE: Differential Gene Expression analysis
Introduction
In spatial biology, the acquisition of spatially resolved omics and deep learning (DL) can pave the way for significant discoveries in cancer. Tumors are composed of diverse cell types that communicate spatially and finding their spatial configurations can potentially enable us to understand how tumor cells reconfigure their spatial distribution, communicate with each other, escape immune surveillance, develop drug-resistance, and eventually metastasize [1]. In this context high-resolution single cell spatially resolved transcriptomics (spRNAseq) provide a new perspective of the tumor microenvironments (TMEs) that promise to answer questions we never even knew could be ask. At present there are several spRNAseq technologies. Here we adopt for our discussion CosMX Spatial Molecular Imager (SMI) platform from Nano string. However, there are several other important technologies like 10X-Xenium. CosMx is the first high-plex in situ analysis platform to provide spatial multiomics with formalin-fixed paraffinembedded (FFPE) and fresh frozen (FF) tissue samples at cellular and subcellular resolution [2]. This technology allows to identify spatially transcripts for 1000 mRNAs at subcellular resolution. Once data for a sample of tissue is processed in the SMI, there are several steps that need to be perform before we can observe the TMEs. The main steps of the pipeline include cell segmentation, clustering and cell typing. Due to the high dimensionality of the data and high levels of noise, DL algorithms are becoming the best choice for data processing of spRNAseq [3].
Materials and Methods
Human NSCLC (RNA) dataset was downloaded from nano string.
Dataset was reprocessed using our custom clustering/cell typing pipeline. Next, we classify different TMEs using the cell type composition of patches where each patch consists of a subset of cells.
Result and Discussion
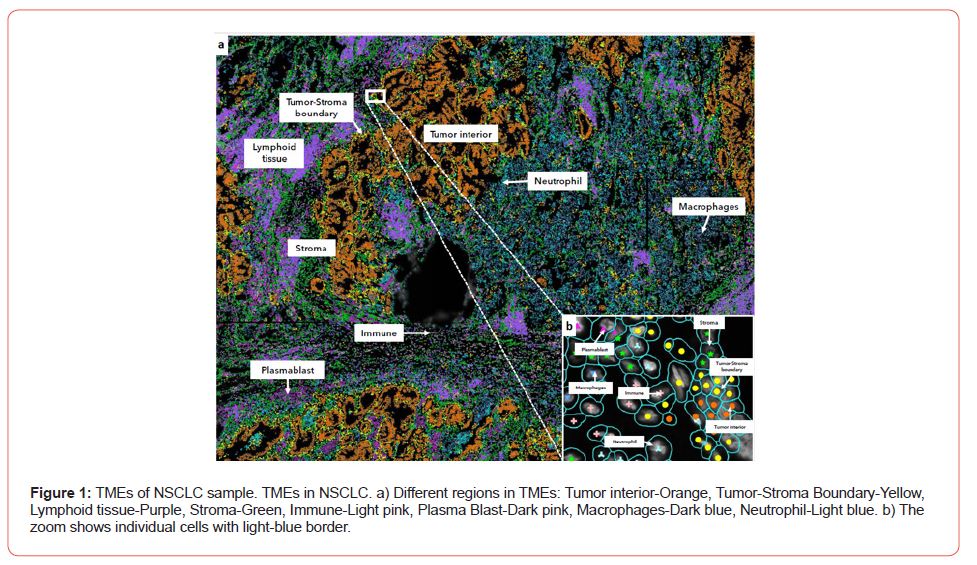
Once cell types are annotated the dataset can be visualized and TMEs categorized (See Figure 1a). Next regions of interest can be obtained to answer specific research questions. Here we present two questions related to TMEs. First question is related to the differential cell and molecular composition of distinct regions within the tumor. Second question builds up by looking at the cell and molecular differences of regions that show immune infiltration.
Differential Cell Distribution and Gene Expression of Distinct Tumor Regions
We refer here to the differences in cell composition as well as in gene expression in contiguous regions of the TMEs. One example is the differential proportion of cell types when we compare the Tumor interior of a region to the Tumor-stroma boundary (See Figure 1a). The statistically significant differences in the proportions of celltypes can be the starting point to additional questions like how heterogeneous the tumor boundary is. Additionally, it is possible to compare the expression of distinct regions of the TMEs and perform Differential Gene Expression (DGE) analysis to identify genes up/down regulated. For example, DGE of Tumor interior vs Tumor-Stroma boundary of the white box (See Figure 1a) shows that gene Transmembrane-4 L-six family member-1 (TM4SF1) is upregulated in tumor-stroma border compared to tumor interior. Interestingly it has been reported that TM4SF1 promotes non-small cell lung cancer proliferation, invasion and chemo-resistance in NSCLC [4] (Figure 1).
Cell and Molecular differences of regions that Show Immune Infiltration
A fundamental aspect of TMEs is immune infiltration [5]. Spatial transcriptomics can be used to study infiltration of immune cells in different regions of the TMEs and at different resolutions (See Figure 1a and 1b). This is important since the colocalization of intracellular transcripts can potentially inform about the signals to which a subset of cells are exposed [6]. Also, it can provide an indirect estimation of the enrichment of pairwise ligand-receptors at the interface of interacting cells.
Conclusions
The use of high-resolution spatial transcriptomics together with its processing with DL are at the fore-front of research of TMEs. These technologies are already expanding our view of the microenvironments. However, there are still a number of challenges related to the quality and integration of datasets, the lack of standardization between different technologies, the cost and the acceptance of the medical community. The future of spRNAseq will continue to increase how much we can see when we study the complex and heterogeneous microenvironments of tumors.
Acknowledgments
None.
Conflict of Interest
No conflict of interest.
References
- Salmen F, Stahl PL, Mollbrink A, Navarro JF, Vickovic S, et al. (2018) Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat Protoc 13(11): 2501-2534.
- He S (2022) High-plex Multiomic Analysis in FFPE at Subcellular Level by Spatial Molecular Imaging. bioRxiv preprint.
- Flores M, Liu Z, Zhang T, Hasib MM, Chiu YC, et al. (2022) Deep learning tackles single-cell analysis-a survey of deep learning for scRNA-seq analysis. Brief Bioinform, 23(1).
- Ye L, Pu C, Tang J, Wang Y, Wang C, et al. (2019) Transmembrane-4 L-six family member-1 (TM4SF1) promotes non-small cell lung cancer proliferation, invasion and chemo-resistance through regulating the DDR1/Akt/ERK-mTOR axis. Respir Res 20(1): 106.
- Sun H, Sui B, Li Y, Yan J, Cao M, et al. (2021) Analysis of the Significance of Immune Cell Infiltration and Prognosis of Non-Small-Cell Lung Cancer by Bioinformatics. J Healthc Eng p.3284186.
- Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A, et al. (2022) An introduction to spatial transcriptomics for biomedical research. Genome Med 14(1): 68.
-
Karla Paniagua, Yufang Jin and Mario Flores*. Looking at the TMEs from the Spatial Transcriptomics Perspective. What can we see?. Adv Can Res & Clinical Imag. 3(5): 2023. ACRCI.MS.ID.000572.
-
Spatial, Transcriptomics, Tumor, Microenvironment, Computational, Visualization, Spatial Molecular Imager, Fresh Frozen, Cancer
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






