 Review Article
Review Article
The Ontology of Tooth-Diseases for Jordanian cases
Riad Jabri, Department of Computer Science, The University of Jordan, Amman, Jordan.
Received Date: February 13, 2020; Published Date: March 20, 2020
Abstract
Ontologies are developed to share the knowledge among the researchers working on the same domain. In medical domain it is becoming an important aspect of medical informatics. The objectives of building medical ontology are constructing more powerful and more interoperable information systems in health care. This paper builds a tooth-diseases ontology that represents the anatomical structure to the diagnosis of diseases. We consider the tooth diseases among Jordanians. Frequent diseases were selected to be represented after discussions with a dentist’s specialist. Their symptoms and respective treatments are then formalized. Finally, OWL based ontology is constructed. Thus, obtaining a formal diagnostic model for tooth diseases. This enables better understanding and diagnosis of such diseases by dentists, dental students and the ones who lack field experiences.
Keywords: Disease; Ontology; Medical; Tooth; Description logic
Introduction
The term “ontology” has its roots in philosophy which has been defined as a particular theory about the nature of being or the kinds of existence. In the computer science community, ontology becomes an important issue. Many research areas study ontology, such as Artificial Intelligence, knowledge-based systems, language engineering, multi-database systems, Medical area, agent-based systems, information systems, etc [1].
Medical ontologies have become prominent in recent years, not only for medical researchers but also physicians, hospitals and insurance companies. Medical ontologies linked disease concepts and properties together in a coherent way. This paper focused on developing tooth-disease ontology which contains diseases of tooth, symptoms, cause, and classification information related to their treatment. In particular, we develop tooth-diseases ontology (TDO) with frequently occurring tooth diseases in Jordan. We identify frequent tooth diseases and formulate their symptoms and treatment, using description and defeasible logics. Protege OWL ontology is then constructed to diagnose an appropriate treatment of the selected diseases. Thus, achieving a better understanding of the tooth diseases for ones who have less experiences. Further, this ontology can be linked to a simulation model for education of dental students by offering the necessary information in their fields.
This paper is structured as follows: section two provides the background and motivation for developing TDO. Section three identifies TDO. Section 4 presents a formalization of TDO. Section 5 presents the construction of TDO. Finally, in Section 6, we draw our conclusions.
Background and Motivation
The lack of deep knowledge in a domain is the major bottleneck preventing the rapid spread in knowledge bases. Nowadays, ontology systems have appealed more and more attention in several research areas such as medical vision. Where a lot of ontologies vision systems have been presented and have achieved great success for handling complicated medical domains. In spite of an ever-increasing number of biomedical ontologies, there are relatively little ontology available for use by the dental community at the present time, one reason for this may be that the potential uses, and applications of dental ontologies have not been adequately described. This paper represents an attempt to address this issue. We choose to work in dental diseases for Jordanians cases as an opportunity to represent these cases. In addition to the fact that Jordan have many distinctive cases in dentistry such as the case found by Haddadin et al. [2] and Faiez N. Hattab [3]. Concisely, our research interest lies on the area of dental community which consequently enriches the semantics of the knowledge to be utilized as knowledge base.
In recent years, many researchers have been carried out to propose new medical ontology, Khoo et al. [4] have state initial version of disease-treatment ontology developed based on an analysis of 50 medical abstracts on colon cancer therapy retrieved from the Medline database.
Information technology today is widely adopted in modern medical practice, especially supporting digitized equipment, administrative tasks, and data management but less has been achieved in the use of computational techniques to exploit the medical information in research. Tokosumi et al. [1] have suggested locality of knowledge for a practical use of ontologies, coordination of large integrated ontologies and localized community ontologies were proposed. Constructing knowledge repository in ontological expressions, that is easy understandable for knowledge agent in the community, enables to maintain safety on the medical communication and to improve the quality of medical care.
In the dental case Park et al. [5] presents a dental case study, calling for reasoning with an OWL-DL ontology and SWRL-based rules to helping decision of restoring a missing tooth. While Kiani et al. [6] proposed system to negotiation between dental experts over the treatment of wisdom teeth. These systems are designed to provide help to the dentist in improving oral health status and to potentially reduce errors in practice.
Jensen et al. [7] develop Neurological Disease Ontology (ND) to provide a framework to enable representation of aspects of neurological diseases that are relevant to their treatment and study. While Ashburner et al. [8] show the success of the Gene Ontology in how a controlled and properly curated ontology can benefit and extend research in medicine. In Scheuermann et al. [9] represent Ontology for General Medical Science demonstrate entities in the domain of medicine and disease and addresses the need to integrate biomedical data. The description logic as we use in this paper is expressive enough for defining the relevant concepts in enough detail, but not too expressive to make reasoning infeasible. The Gomes et al.[10] describe structure of description logics and its approach in the cardiology medical environment, and we compare the use of description logics in the pathology environment by using a practical model of description logics use in terms of diseases related to the circulatory system of the human body.
Tooth- Disease Ontology
Several methodologies for constructing ontology have been suggested [11]. In this section, we build tooth- disease ontology (TDO) following a well-accepted method [11] as shown in (Figure 1).
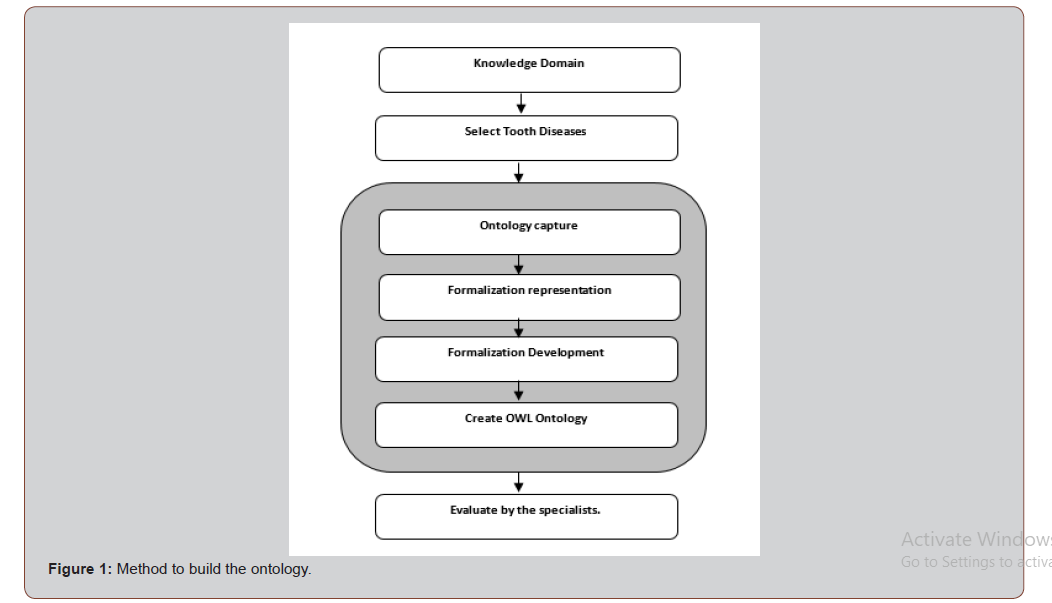
In the following subsections, we will address each step of TDO building method.
Knowledge domain
We define the domain of TDO as consisting of tooth diseases, symptoms, and their respective treatments. TDO is new one, where there is not much research done on it, especially in Jordan. Thus, we define our scope as to develop TDO in reusable form of knowledge. Dentists, especially the ones who lack experiences, will be able to get help for their decision making and to learn what terms in the domain mean.
Selecting tooth diseases
The tooth diseases were selected based on the ones that are classified as a highly occurring among Jordanian, as shown in Table I, The terms used to express these dieses and TDO are the same as the ones used in medical dictionaries and schools for education (Table 1).
Table 1: Selected Diseases.
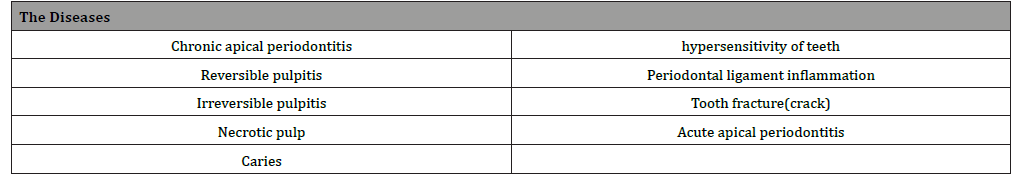
Ontology capture
Building the ontology is reduced to identification of the key concepts and their respective relationships. Thus, the top-level classes or concepts of TDO are shown in Figure 2. Each node represents a class or concept. Each directed arc or arrow represents a property or relation. The top-class in the ontology is TDO which represents specific treatments that are considered for a particular disease, as described in the paper (Figure 2).
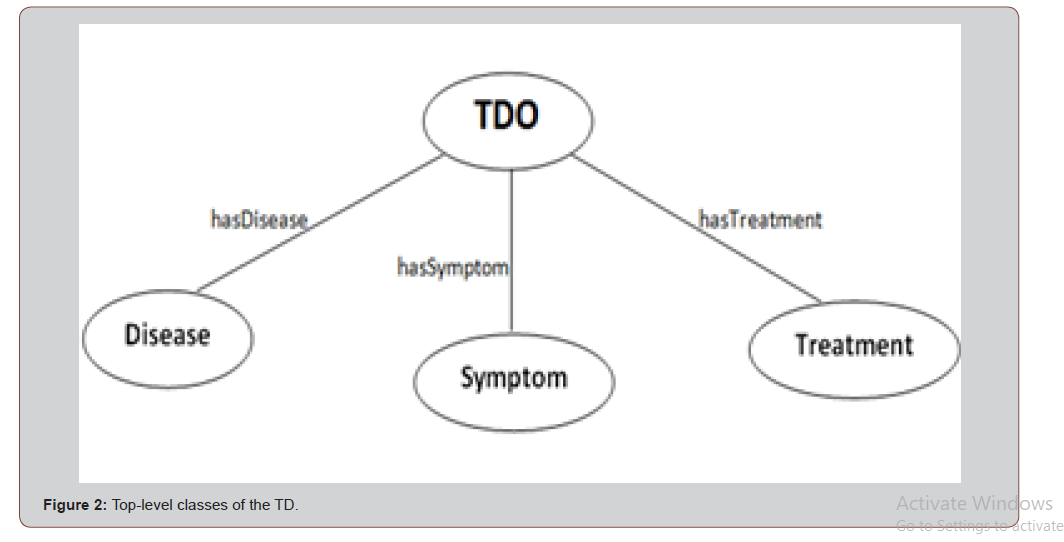
Information related to a disease-treatment is divided into three aspects, represented by the following properties and top-level classes:
• Has Disease-> Disease
• Has Treatment-> Treatment
• Has Symptom-> Symptom
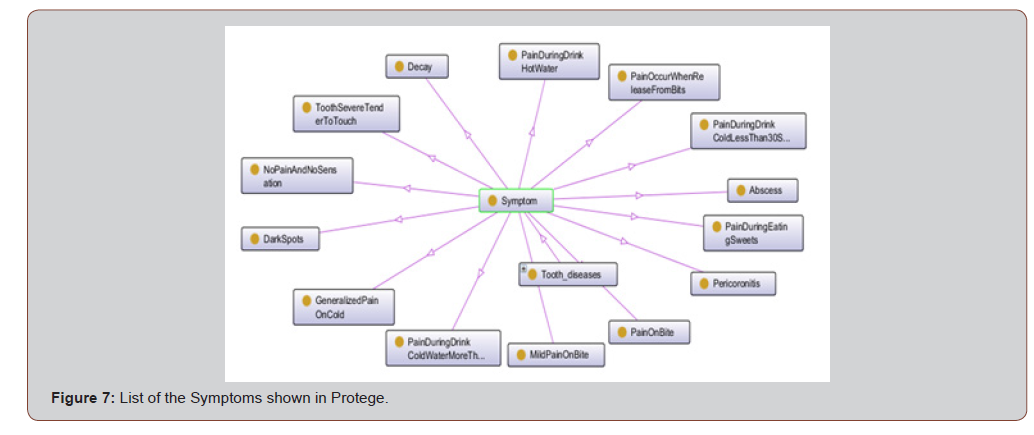
The Symptoms class is the superclass of 14 subclasses which are representations of different symptoms in the domain, for examples: pain with sweets, pain while drink cold water, etc. Figure 2 shows some of these subclasses. All the subclasses of Symptoms are common or global concepts, vocabulary, between individuals (Figure 3).
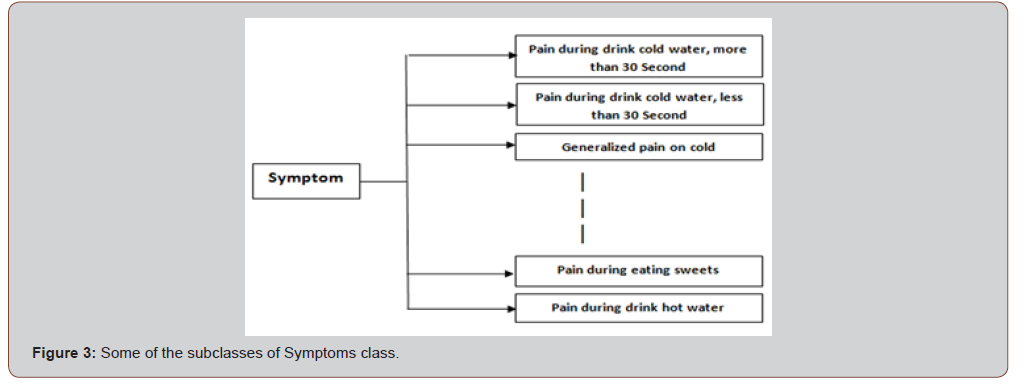
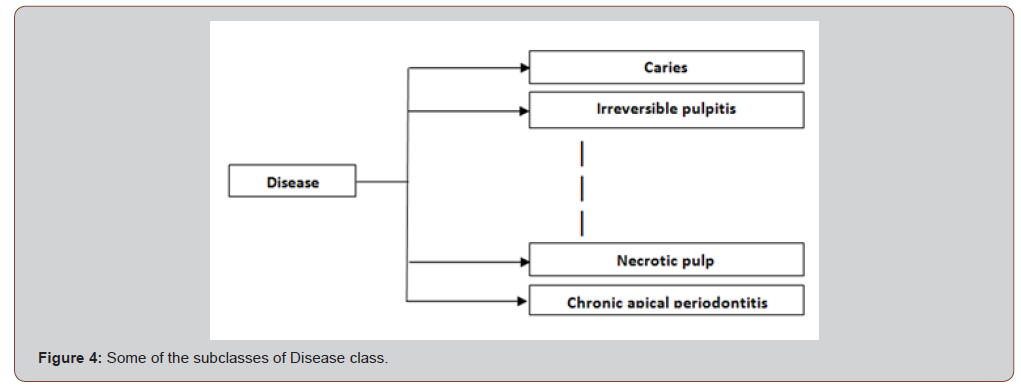
The Diseases class is the superclass of 9 subclasses which are representations of different symptoms in the domain, for examples: Caries, Irreversible Pulpitis, etc. Figure 4 shows some of these subclasses (Figure 4).
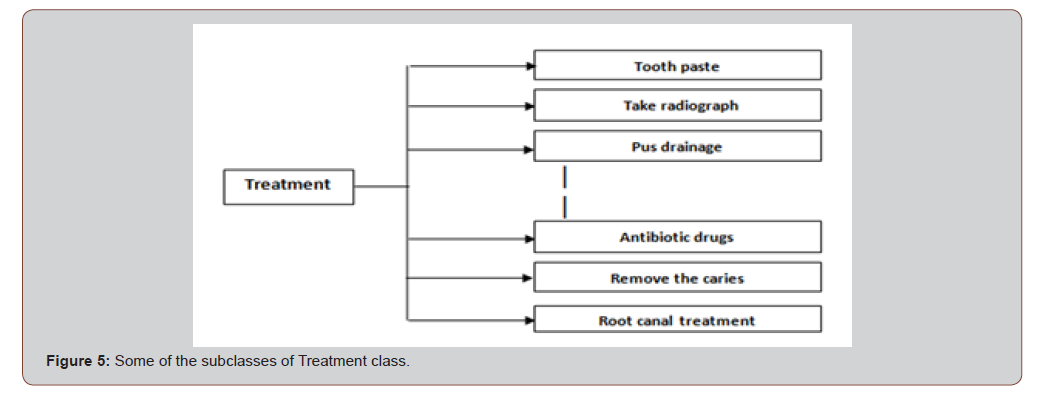
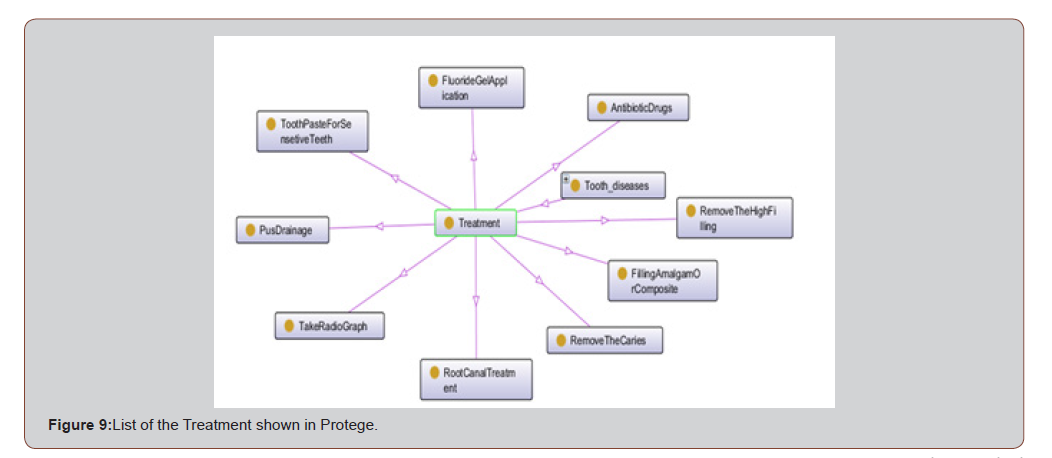
The Treatment class is the superclass of 9 subclasses which are representations of different symptoms in the domain, for examples: Antibiotic Drugs, Root Canal Treatment, etc. Figure 4 shows some of these subclasses (Figure 5).
Formal Representation
The Basic ontological can be formulated of assertions concerning the entities in a domain by using logical resources; we use the family of Description Logics (DLs) [12], Combined with Defeasible reasoning [13,14].
Description Logics are subsets of first-order logic. Although DLs are far from being able to express everything, it is important in providing a standard for representing domain knowledge (logical formalism for ontologies) in the context of the Web Semantics; the Web Ontology Language (OWL) is based on Description Logics. And the DLs are supported by many tools, such as the Protege editor. But the main important about DLs that have certain favorable computational properties because is decidable which means that algorithms exist for which it is guaranteed that they will always return some result. On the other hand, Classical reasoning for logic-based Knowledge Representation systems is in general, monotonic like the DLs, where the assumption in these systems that there is complete information about a domain. This means that they generally cannot deal with any new information arising which contradicts with the current information. But in the medical field and specifically in the disease diagnosis process, we need non-monotonic systems that have been investigated which can reason under uncertainty or within complete information. Defeasible reasoning is one particular model for implementing non-monotonic reasoning [13,14].
DL based formalization
This section illustrates the use of description logics for the tooth diseases ontology; Thus, with well-structured and well-defined description logic structure, knowledge base, terminologies, and its object inference are precisely defined.
In the following, we illustrate the DL syntax and semantics through a set of examples, where:
• Entity names starts with upper case letters, e.g.
Periodontal_Ligament_Inflammation.
• Relation symbols start with small case letters, e.g. has_ Symptom.
• Roles starts with a quantifier, followed by a relation symbol, followed by a dot and an entity symbol
Example 1: Formalization of diseases symptoms
• Symptoms for Caries

Example2: Formalization of diseases treatment
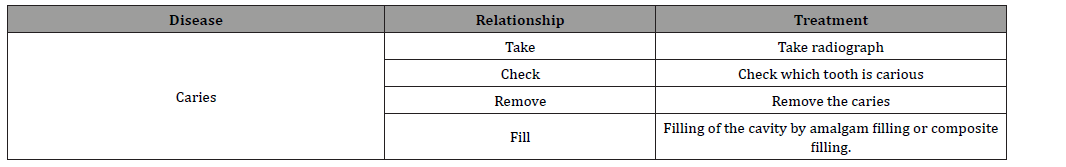
Treatment of Caries Table 2 shows the entire treatment and relationships respective to Caries. We formulize the treatment Remove caries by the following expression
Table 2: Entire Treatment and Relationships for the Entity “Caries”.

∃ has Method. Removal Action denotes the entity whose instantiation is the set of all individuals related to an instance of Removal Action by the relation has Method.
expression:

Defeasible logic formalization
In defeasible logic , the strict rules are assigned that a fact is constantly a consequence of another ;Whenever the conditions are incontrovertible then so is the conclusion, , defeasible rules that can be defeated by contrary evidence; and Superiority relations among rules are used to define priority ordering over the defeasible rules and the defeaters can be given. Such rules are used to express treatment exceptions as illustrated by the following example.
Example 3: Pregnant exceptions
The treatment process of tooth disease may have a radiograph to check of something this step is necessary to check of caries or something else during the treatment procedure. But if we take the pregnant women as example in this case this procedure must exclude because the radiograph very danger of the baby. This case can represent it by the defeasible logic, so we can represent the knowledge base depend on Moodley et.al.[13] as:

This means that when a patient comes to the doctor suffering from Tooth pain, it will be very important to ask the patient about the pregnancy to know if they can use the radiograph or not. Otherwise, the patient can be doing all the procedures they need to the right treatment.
Building OWL ontology
OWL is used mostly with ontology language. It is a W3C recommendation to be “used by applications that need to process the content of information instead of just presenting information to humans” [15]. It has several features over XML and RDF by providing additional vocabulary along maintain their properties. It is then used to define individuals and maintain their properties, and then it is used to reason about these classes and instances.
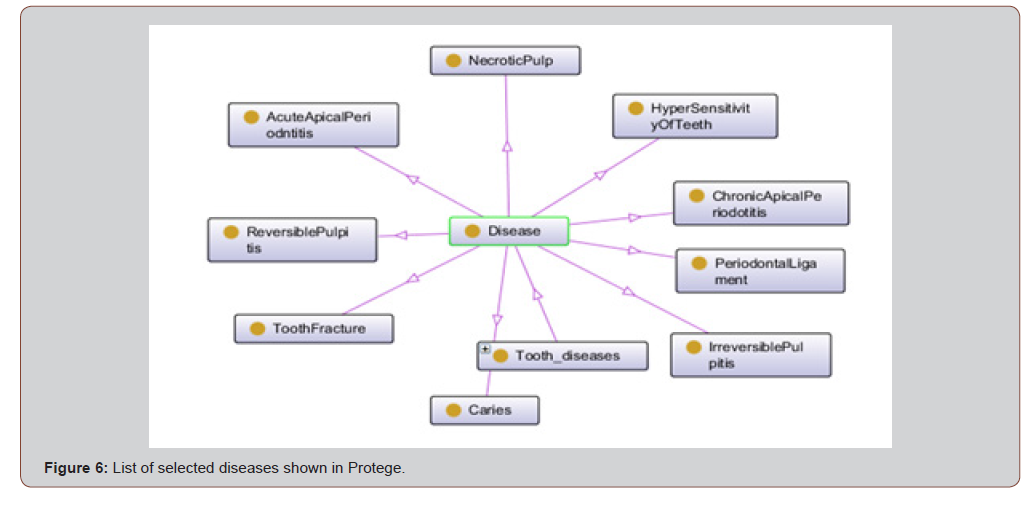
Figure 6 presents Protege OWL ontology graph created from the tooth ontology of the nine selected diseases. ⟨Disease⟩ was defined as a superclass and the 9 selected diseases were treated as classes. [Figure 6]
While figure 7 presents Protege OWL ontology graph created from the Tooth ontology of group of symptoms. ⟨Symptom⟩ was defined as a super class and the 14 Symptoms were treated as classes. [Figure 7]
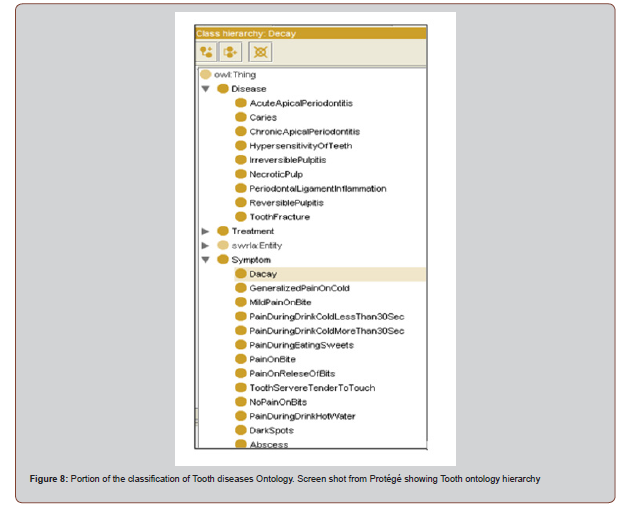
In particular, we have built a large hierarchy of 23 subclasses of ‘Tooth disease’ as illustrated in below Figure. [Figure 8]
Figure 9 presents Protege OWL ontology graph created from the Tooth ontology of group of treatments. ⟨Treatment⟩ was defined as a super class and the 9 treatments were treated as classes. [Figure 9]
To construct TDO, four languages have been used for implementation: Protege, OWL-DL, SWRL, and Java. The ontology of the concepts has been implemented using Protege. Protege is a free open source platform provides users community with a tool to construct domain models and knowledge-based applications with ontologies. OWL-DL is designed to provide the maximum expressiveness possible of OWL while retaining computational completeness along with a formal semantics. OWL DL includes all OWL language constructs, but they can be used only under certain restrictions.
Rules for mapping local concepts to global concepts have been coded using SWRL. SWRL is a Semantic Web Rule Language. Hornlike rules can be defined by the SWRL using the terms of OWL concepts. SWRL is fully compatible with OWL. A reasoner for finding agreements and disagreements has been implemented using Java. [Figure 10]
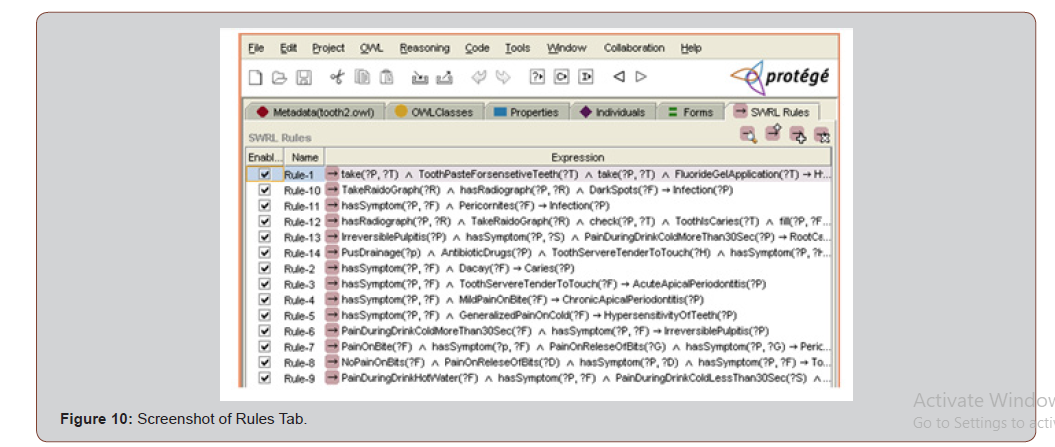
After creating all the properties of the ontology, rules should be executed and new inferred facts (if they exist) should be added to the OWL knowledge base. This step was done using the SWRL and Jess tabs in Protege. Figure 10 is the screenshot of the Protege Rules tab.
Conclusion
We built the ontology of tooth diseases with links to the symptoms, method of treatment of highly occurring tooth disease among Jordanians. If cases are categorized in the reusable form of knowledge, dentists, especially the ones who lack experiences, will be able to get help for their decision making. As future work, this ontology can be linked to a simulation model for education of dental students.
Acknowledgment
None.
Conflicts of Interest
No conflicts of interest.
References
- N Guarino (1998) Formal ontology and information systems. In Proc Of the 1st International Conference on FOIS, Trento, Italy.
- Paulo Gomes, Karina Loncarovich (2011) Description Logics as information resource: as example of its application in cardiology. in RECIIS-R Eletr by Com Inf Inov Health Rio de Janeiro 5(1): 104-116.
- Faiez N Hattab (2014) Double talon cusps on supernumerary tooth fused to maxillary central incisor: Review of literature and report of case 6(4): e400-e700.
- CG Khoo, J Na, V Wang, S Chan (2011) Developing an Ontology for Encoding Disease Treatment Information in Medical Abstract. DESIDOC Journal of Library & Information Technology 31(2): 103-115.
- Tokosumi, Naoko Matsumoto, Hajime Murai (2007) Medical Ontologies as a Knowledge Repository. proceedings of the 2007 IEEE/ICME, Beijing, pp. 487-490.
- Park SG, HG Kim (2006) Dental Decision Making on Missing Tooth Represented in an Ontology and Rules. in Lecture Notes in Computer Science, The Semantic Web-ASWC 2006, Heidelberg: Springer Berlin 4185: 322-328.
- Mahsa Kiani, Michael Francis, Yassaman Zand-Moghaddam, Palash Verma (2010) Chapter 4 Ontology-Based Negotiation of Dental Therapy Options. TMRF e-Book Advances in Semantic Computing, (Eds.) Joshi, Boley, Akerkar 2: 52-78.
- Mark Jensen, Alexander P Cox, Naveed Chaudhry, Marcus Ng, Donat Sule, et al. (2013) The neurological disease ontology. Journal of Biomedical Semantics 4(1):42.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25-29.
- Scheuermann RH, Ceusters W, Smith B (2009) Toward an ontological treatment of disease and diagnosis. Summit on Translat Bioinforma 2009: 116–120.
- Gawich M, Badr A, Hegazy A, Ismail H (2012) A methodology for ontology building. International Journal of Computer Applications 56(2): 39-45.
- Baader F, Calvanese D, Mcguinness Dl, Nardi D, Patel-Schneider PF (2007) The Description Logic Handbook Theory, Implementation, and Applications. 2nd Cambridge: Cambridge University Press, US.
- Kody Moodley, Thomas Meyer, Ivan José Varzinczak (2012) A defeasible reasoning approach for description logic ontologies. 69-78.
- Ronald J (2004) Brachman and Hector J. Levesque, Knowledge Representation and Reasoning, Morgan Kaufmann Publishers, San Francisco, CA, USA.
- http://www.w3.org/TR/owl-features/.
-
Riad Jabri, Enas Faisal . The Ontology of Tooth-Diseases for Jordanian cases. Arch Biomed Eng & Biotechnol. 3(5): 2020. ABEB. MS.ID.000579.
-
Intracellular, Extracellular ice crystals, Cumulus cells, Denuded oocytes, Cryopreservation, Tissues, Organs, Oocyte maturation, Meiotic arrest, Meiotic resumption
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






