 Mini Review
Mini Review
Role of CRISPR-Cas/9 in Hematological Disorders: A Review
Seema Rani Padhiary1, Sameer Sharma1*
Department of BioNome, BioNome Private Limited, Bangalore, India
Sameer Sharma, Department of Bioinformatics, BioNome Private Limited, Bangalore, India.
Received Date: April 20, 2020; Published Date: May 20, 2021
Abstract
The ongoing research in genome editing and gene silencing methods have paved a number of ways in not just detecting the diseases but also in rectifying it too. CRISPR Cas9 technology has been a gift in the field of genome editing technology. CRISPR Cas9 not only helps in identifying the diseases located in a particular gene but also helps in correcting the gene in its genome level by either silencing the gene for generations or replacing the defective gene and modifying it. With the help of CRISPR Cas9 replacing concept the defective gene is replaced with the new one and the disorder is not continued further. This advancement in the technology is the key benefit for the CRISPR Cas9 procedure. To add with it zincfinger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs)are the other two technology to contribute in genome editing too. Haematological disorders being lethal in nature have found a cure via CRISPR Cas9 technology.
Keywords:Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated protein 9 (Cas9) system; Haematological disorders; Gene editing; Haematological malignancies & monogenic
Mini Review
Over the past two decades, advances in molecular biology and genetics have greatly extended our knowledge of hematological diseases. Every year hematological disease are affecting million people from malignancies to acquired coaglupathiesis. However, the characterization of genes of interest remains a great challenge. The optimal way to unravel the roles of particular genes is to manipulate their functions via gene deletion, insertion or modification. Genome engineering is defined as the deliberate modification of an organism’s genetic material. From the 1980s several studies unravel the genetic mutation and their effect on human disease to determine the malignancies and advance diagnosis, prevention and diagnosis. Earlier gene editing technology were homologous recombination which was less efficient, time consuming & laborintensive process. Later in subsequent years, Zinc finger nucleases [ZFNs] & transcription activator-like effector nucleases [TALENs], was introduced to stimulate cellular DNA repair pathways. But there are so many limitations, so to overcome those limitation CRIPR associated CAS9 protein has introduce to start a new era of genome editing. From past few years CRISPR have change the path of genome editing be developing plant and animal models and the alleviation of human disease like hematological disorders. According to research crRNA sequence have significance role in targeting DNA [1,2]. To established CRISPR/Cas9 Jinek et al, 2012 introduce crRNA and tracrRNA into a single 100 nucleotide and a guider RNA which helps in the breaking of double stranded DNA and thus successfully genome editing done in human and mouse cells [3]. In the hematopoietic setting, CRISPR/Cas9 gene editing has been applied both in research and in clinical translation studies. CRISPER/Cas9 can be used to treat the clonal hematopoiesis by recapitulate the genetic mutation in patient. The main goal is to restore the immune system by the next generation genomics studies coupled with CRISPR/Cas9. Several companies develops their therapeutics against auto immune diseases and multiple myeloma by using this editing tools. The question marks comes whether this technique is safe or not, for this clinical trials are ongoing. The basic principle of CRISPR/Cas9 mediate gene modification are 1. Cas9 protein with the help of tracrRNA binds with specific DNA target which is marked by guider RNA with crRNA sequence. 2. Cas9 have endonuclease activity that helps in cleave of DNA. 3. The cleavage site will be repaired by the nonhomologous end joining (NHEJ) DNA repair pathway, by this process insertion/deletion can be done to make the gene functional or dysfunctional.
CRISPR Cas/9: A Multifaceted tool for Genome editing
Clustered regularly interspersed short palindromic repeats (CRISPRs) were originally discovered in E. coli in 1987 [2,4,5]. The CRISPR‐Cas systems are divided into two classes based on the structural variation of the Cas genes and their organization style[2] Specifically class 1 CRISPR–Cas systems consist of multiprotein effector complexes, whereas class 2 systems comprise only a single effector protein; altogether, six CRISPR-Cas types and at least 29 subtypes have been reported [6,7], and the list is rapidly expanding. Type II CRISPR system is used in most of the cases which is depend on single CAS protein developd fron streptococcus targeting DNA and therefore active the gene editing tools [8] Mechanistically, the CRISPR/Cas9 system comprises two components, a singlestranded guide RNA [sgRNA] and a Cas9 endonuclease. To target the DNA need specific target sequence, thus the sgRNA consist of 20 BP and this must be compatible with Cas9 protein used, which is termed the “protospacer adjacent motif” [PAM] of an “NGG” or “NAG” [9,10]. The sgRNA binds to the target sequence by Watson– Crick base pairing and Cas9 precisely cleaves the DNA to generate a DSB [1]. Following the DSB, DNA-DSB repair mechanisms initiate genome repair. Targeted genomic modification can be done with CRISPR/Cas9 through NHEJ or high fidelity HDR [11]. On the other way CISPR/Cas9 with guider RNA have more advantages then other editing system [12]. Endonuclease based ZFN or TALEN tools need to be reengineering for each case of targeting site, however the Cas9 also be engineered to recognized new site via changing the guider RNA sequence. Another one advantages of CRISPR is that it has potential to multiple loci editing by changing the sequence which make its more suitable then other modification system (Figure 1).
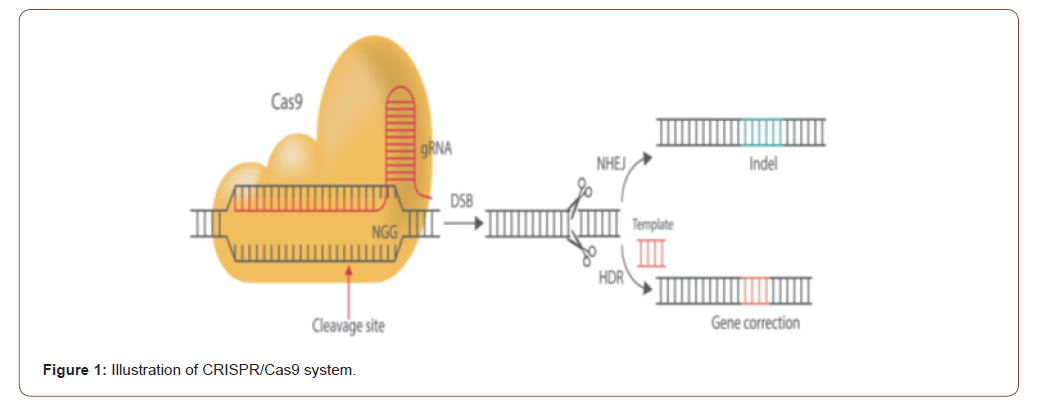
CRISPR/Cas9 is now an indispensable tool in biological research. Three common strategies have been developed for genome editing with the CRISPR/Cas9 platform (1) The plasmid‐based CRISPR/ Cas9 strategy, where a plasmid is used to encode Cas9 protein and sgRNA, [3,13] assembles Cas9 gene as well as sgRNA into the same plasmid in vitro. This strategy is longer lasting in the expression of Cas9 and sgRNA, and it prevents multiple transfections [14]. However, the plasmid needs to be introduce inside the nuclease of target cells.
The ethical and security risks comes under the consideration when gene editing done because it is irreversible. Some times its causes complicated nerves disease. As result genome editing can be done only for RNA modification by proposal of scientist. The byproduct of transcription, RNA is responsible for downstream processing of protein. RNA modification is one of the best methods to treat the disease to avoid the irreversible modification and mutation. In therapeutic genetics CRISPR can be introduce with the stem cell therapy. Pluripotent stem cells have ability to differentiate in retinal precursors, and it has been proven that CRIS with stem cells have less immune rejection problems [19.20]. However, patient-derived iPSCs might still harbor the same pathogenic genes, which could influence the therapeutic efficacy of transplanted cells.
ITherefore, it is necessary to combine the CRISPR/Cas9 system to fix disease- causing mutations in patient derived iPSCs before transplantation [21]. CRISPR was first described in Escherichia coli as an odd 29- nucleotide repeat sequence with 32-nucleotide spacing by a Japanese research group in 1987, although no further detailed observation was reported [22]. Several years later, Francisco Mojica discovered similar repeats in several microbial species and reported their potential use in prokaryotes [23]. By 2002, the novel repeats were formally named as Clustered Regularly Interspaced Short Palindromic Repeats [CRISPR] meanwhile, scientists discovered specific CRISPR-associated (Cas) genes that were closely related to the function of CRISPR [24]. In 2003, Mojica discovered the function of the CRISPR system, proposing that CRISPR is an adaptive immune system that protects microbes against specific infections [23]. Two independent groups [4,5] reported similar conclusions, and their finding was confirmed by experiments showing that CRISPR provides acquired immunity against viruses in prokaryotes [25]. Researchers from the Massachusetts Institute of Technology have developed and optimized the Streptococcus pyogenes CRISPR-Cas9 system to facilitate efficient genome editing in mammalian cells [3]. Another group established a CRISPR-Cas9 system as an RNAguided editing tool for human genome engineering using a chimeric single-guide RNA [sgRNA] instead of a tracrRNA:crRNA duplex.
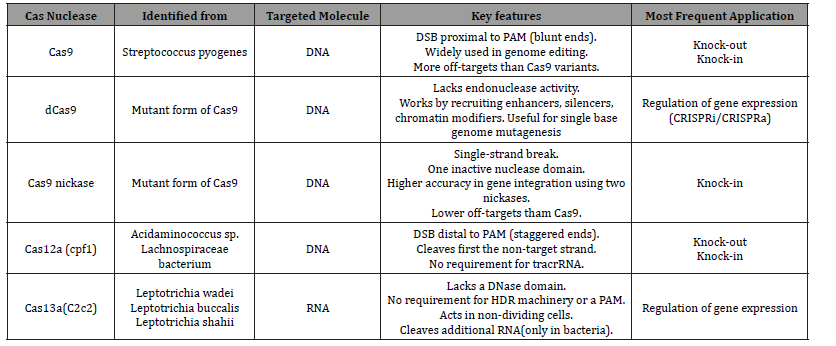
A CRISPR-Cas9 system was applied to screen for loss-offunction using a high-throughput screening platform in human cells [26]. These genome-scale knockout screens provide an alternative screening system to RNA interference (RNAi), which is limited by partial depletion of target gene levels, toxicity and transient silencing [27]. [28] A catalytically inactive Cas9 protein [dCas9] was demonstrated lacking endonuclease activity that can be used as a platform for RNA-guided transcription regulation; this modified system is called CRISPR interference [Crispi]. Unlike RNAi-based silencing, which destructs transcribed mRNAs, the CRISPR-dCas9 system directly blocks transcription elongation within proteincoding regions and leads to dramatic suppression of transcription, with no detectable off-target effects [28]. The authors subsequently reported that both repressive and activating effector domains, such as KRAB and VP64, can be fused to dCas9 to either repress [CRISPRi] or activate [CRISPRa] the transcription of target genes [29]. Other than targeting dsDNAs, CRISPR-Cas9 was modified to bind or cut RNA targets in vitro by providing the proto spacer adjacent motif [PAM] as part of an oligonucleotide [PAMmer]. RNA-targeting Cas9 [RCas9] as a tool to track RNAs in living cells, in which RCas9 is capable of tracking the localization and movement of endogenous RNAs (Table 1).
Table 1: Comparison of the most widely used Cas nucleases.
For treating the hematopoietic cells, the main problem is efficiency of nontoxic delivery of components into therapeutic relevant cells like stem cells and T cells. However, the balancing need to be required the CAS9 RNP act in duration as short as possible. According to the several studies prolonged expression leads to higher chances of unintended activity at closely match target sequence. In general, there are different methods to deliver the CRISPR/Cas9 in the system.
• The 1st method DNA plasmid-based CRISPR/Cas9 system, which encodes the Cas9 protein and sgRNA from the same plasmid.
• The 2nd method ‘all RNA’-based delivery method which Cas9-encoding mRNA and the sgRNA are delivered simultaneously.
• The last approach is delivery as an RNP complex consisting of recombinant Cas9 protein complexed to the sgRNA prior to delivery. Transfection with a plasmid-based CRISPR/Cas9 system has been extensively used in easy-to-manipulate cells. However, the major problem with this system is the induction of an innate immune response to foreign DNA, which is lacking in many cancer cell lines. A forementioned plasmid delivery in haematopoietic cells increases the cellular toxicities [118,119]. Electroporation with RNP complexes or Cas9 mRNA and sgRNA have viewed to work very systematic in both the cells along with RNP system giving slightly higher INDEL frequencies [119,120,121]. Mordern transcriptomic profiling after Cas9 mRNA transfer also suggests that mRNA does, to some extent, invoke an innate immune response in HSCs, which is not the case for Cas9 RNP delivery [122]. One limitation of this delivery modality is the apparent instability of the sgRNA, which leads to relatively low gene editing rates, particularly in primary cells [119].
Applications of Crispr/Cas9 in Haematological Disorders
The simplest genome editing system is type II CRISPR tool which require Cas9 protein and tracrRNA and crRNA [2,30]. This duplex is simplified by sgRNA and Cas9 binds with double standard DNA and cleave it by endonuclease activity. Thus, it trigger NHEJ or HDR mediated genome editing [2]. The functions and mechanisms of the type II CRISPR-Cas9 system have been extensively reviewed in detail [31-33].
Non-cancerous haematological disorders
Beta-thalassaemia
b-thelassaemia caused by mutation in the human hemoglobin beta [HBB,also known as b- globin] and this is most common inherited blood disorders. The decreasing expression of HBB resulting erythropoiesis and thus severe anemia [34,35]. To treat this disorderd allogenicHSCT therapy is present previously but in resent era CRISPR Cas9 mediated gene editing is used to correct the HBB gene mutation via HDR. Apart from that stem cell also introduced to treat this erythropoiesis [37-39]. Gene-corrected iPSCs can restore HBB expression with a minimal off-target effect. In a recent study, the fibroblasts of a b-thalassaemia patient were reprogrammed to become transgene-free natıve-state iPSCs, which showed significantly higher targeting efficiencies in the CRISPRCas9 genome editing system compared with primed iPSCs [40,41] Unfortunately, therapy via HbF reactivation has lagged for many years due to the unclear regulatory relationship between HbF and adult haemoglobin [HbA] during the development of erythroid cells. Recently [42] identified an erythroid-enhancer region within the BCL11A gene via CRISPR-Cas9-mediated in situ saturating mutagenesis. As an ideal therapeutic target for b-haemoglobin disorders, this region can be disrupted by individual sgRNAs, leading to efficient HbF reactivation.
Sickle-cell disease
SCD is another genetic disorder caused by deficiency of oxygen [43] it affects the red blood cells and form a sickle shape, clogs blood vessels and shows pain full symptoms like such as haemolysis, ischaemia, anaemia and multi-organ injury [44,45]. Several research attempts to correct the mutation by ZFN and TALEN induced HDR with the help of stem cells [46,47]. One recent study utilized a CRISPR-Cas9 system to target the endogenous HBB locus in human iPSCs generated from SCD patients, resulting in higher efficiencies compared with other nucleases [48]. In addition BCL11 gene targeting by CRISPR-Cas9 also can used to treat this [49].
Thrombocytopenia
Thrombocytopenia is an inherited or acquired alteration of genes, its lead to decreasing of thrombocytes [50]. Till now only few cases are documented as CRISPR treatment [51]. According to recent studies CRISPR-Cas9 gene editing to convert megakaryocytelike cells and iPSCs from HPA-1a (human platelet alloantigen-1a) to the HPA-1b alloantigenic epitope, it’s a tough technique.
Haemophilia
Hemophelia is one of the common hereditary disorders of blood coagulation [52]. Hemophilia A caused by in the deficiency of factor viii and hemophilia B caused by deficiency of factor IX. There are several approaches done through Adenovirus-associated virus [AAV] vectors expressing to fix this [53,54]. Because AAV-mediated gene transfer is applicable only to a limited group of patients, alternative strategies have been proposed [High, 2012]. Last year, one research group developed a CRISPR-Cas9 system to revert two inverted chromosomal regions back to the normal orientation in HA patient-derived iPSCs.
Malignant haematological disorders
Myeloma
Worlds second most hematological disorder is multiple myeloma according to NHLs it happens because of malignant plasma cells in the bone marrow. According to research TALENs are used to treat MM [55], utilized CRISPR-Cas9 editing to silence MUC1-C, an oncogenic trans-membrane protein, and found that MUC1-C occupies the MYC promoter and activates the MYC gene via a b-catenin/TCF4-mediated mechanism. Further analysis using CRISPR-Cas9-mediated microarray datasets demonstrated that MUC1-C drives MYC expression and contributes to MM progression.
Leukaemia
In myelod lukaemia CRISPR editing has been overwhelmingly application [56]. Used the CRISPR-Cas9 system to generate mouse models of AML using lentivirus-mediated editing of multiple genes [e.g., Runx1 and Tp53, among others] in murine haematopoietic stem and progenitor cells [HSPC]. This work highlights the application of CRISPRCas9 to generate ex vivo genome editing to delineate the complexity of human blood cancers. Later that year [57]. discovered a novel tumour suppressor gene, KMT2C [lysine methyltransferase 2C, also termed MLL3], in chromosome 7q using an ex vivo CRISPR-Cas9-mediated approach. Further experiments in a mouse model confirmed that KMT2C is a haplo insufficient tumour suppressor in AML and its inhibition impairs differentiation of the myeloid lineage and produces an MDS-like syndrome in mice. Interestingly, murine AMLs with KMT2C suppression are resistant to conventional chemotherapy. This study provides a valuable clue for the treatment of AML patients harbouring 7/del[7q] lesions.
Secondary haematological disorders
Human immunodeficiency virus (HIV) infection
HIV infection has been associated with a broad range of haematological disorders [58]. ZFN-induced disruption of the CCR5 gene, an essential HIV-1 co-receptor required for virus entry, had become a promising gene therapy procedure [59]. CRISPR-Cas9 have further transformed the studies of HIV-1 [60]. DNA division activity of a CRISPR-Cas9 system for silencing the CCR5 gene in vitro, have reported CRISPRCas9- mediated disruption of CCR5 [61,62] and other co-factors, including CXCR4 [63] and XPO1 exportin 1 [64]. The removable of pro-viral HIV-1 DNA from the host cell genome, have reported the potential of the CRISPR-Cas9 system to mutations by targeting the long terminal repeat [LTR] region [65- 68]. Moreover, the feasibility of HIV therapy has been questioned by the latest finding that viewed viral escape from CRISPR geneediting attack due to the Cas9/ sgRNA-derived [69]. CRISPR-dCas9- derived regeneration of HIV-1 is specific in all latency models and does not induce off-target effects [70] (Figure 2).
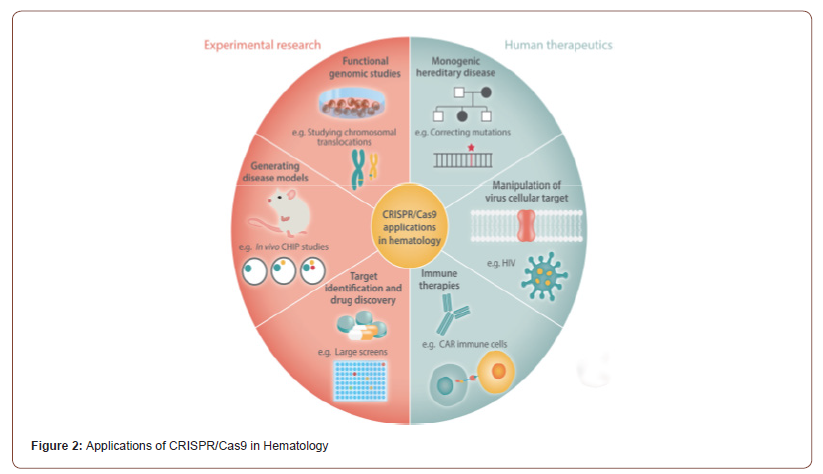
Applications of CRISPR/Cas9 in Hematology
Malaria
Since the CRISPR-Cas9 was successfully applied in Drosophila in vivo [71]. scientists have attempted to apply to target either malaria parasites [72,73] or mosquitoes [71,72]. In a representative study, it was identified that the gene required for fertility in female Anopheles mosquitoes can be inserted via CRISPR-Cas9 gene-drive construct and targeting the loci [74]. This procedure made a kind of sterility in female mosquitoes, paving a way to extinction of the population. Parasite transmission was baffled by incorporating into an anti-malarial gene into malaria mosquitoes via the help of CRISPR-Cas9 [71]. This strategy however declined the graph of malaria but have not lead the diseases to extinct [75,76].
Technical restrictions
CRISPR-Cas9-mediated gene enhancement can be gained by transient or stable transfer of the CRISPR components [3,13,60], Viral-based transfection of CRISPR components is a more efficient substitute to create stably modified cells [56,77,78,79]. [80] Development of rapid cell mechanical deformation has been developed which creates transient holes in the membrane that helps in the transfer of biomaterials through the medium. The MCL1, an anti-apoptotic BCL2 family member, is required for the sustained survival of human BL cell lines because silencing of MCL1 leads to rapid cell death. In this case, the drug-inducible CRISPRCas9 lentiviral system provides an option to identify the functions of tumour-essential genes [81]. Due to the increased utilization of the NHEJ pathway over HDR in mammalian cells, the low efficiency of CRISPRCas9- induced HDR is an additional limitation [13,82]. Therefore, increasing the efficiency of HDR facilitates the rapid generation of cell lines with precisely edited genes. To resolve this issue, a recent study identified a RAD51-stimulatory compound, RS-1, which enhances the efficiency of Cas9-stimulated HDR by 3- to 6-fold [83].
Off-target effects
CRISPR-Cas9 in mammalian cells, have promising results but the question of specificity is still of major concern [84-88]. Deep sequencing [85,86] and whole genome sequencing [89,90] have been considered options to identify mismatches however, the possibility of overlooking off-target sites and the higher cost make it difficult to perform this step in every experiment. Alternatively, a series of online bioinformatics tools were developed to predict off-target mutation sites [11,92-94]. Genome-wide Unbiased recognition of DSBs authorize by Sequencing (GUIDE-seq) provides an unbiased and genome-wide method for identifying CRISPR RNA-guided DSBs in cells [91,95,96]. These studies have enlarged the technical feasibility of off-target detection [97,98], although ‘it is impossible to draw broad conclusions about the specificity of engineered nucleases’, as recently reported [99,100] (Table 2).
Table 2: Current clinical trials in hematology using the CRISPR/Cas9 system.

Consequences and Future Perspectives
CRISPR-technologies have been flourished beyond genome editing while simultaneously research is going on, either NHEJ or HR using then conventional CRISPR/Cas9 platform. The superiority of the procedure is that it helps in converting of one base pair with another without instigating any DSBs and without depending on the HR pathway or systematic delivery of a DNA donor template. The vivo HSC gene therapy with retroviral vectors has given a promising result with [101-103], CRISPR/Cas9 gene editing in a mouse model of b-thalassaemia also [104]. The pre-existing immunity in homo sapiens is potentially an obstacle Cas9 in humans [105,106], which may be avoided by administration of immunosuppressive drugs [107]. CRISPR Cas9 has shown positive results in the field of genome editing and complete gene silencing technology. The utility of this technology in the field of haematological disorders have saved a number of lives. CRISPR Cas9 is a modern technique which uses bioinformatic tools to locate the mutated gene, and to top it even helps in rectifying it too. The use of this technology in curing haematopeotic cells are very promising and have a wider prospective in future too, to deal with any clinical manifestations.
Acknowledgement
None.
Conflicts of Interest
No conflicts of interest.
References
- Hsieh LE, J Sidney, JC Burns, DL Boyle, GS Firestein, et al. (2021) IgG Epitopes Processed and Presented by IgG (+) B Cells Induce Suppression by Human Thymic-Derived Regulatory T Cells. J Immunol 206(6): 1194-1203.
- Schulten V, L Westernberg, G Birrueta, J Sidney, S Paul, et al. (2018) Allergen and Epitope Targets of Mouse-Specific T Cell Responses in Allergy and Asthma. Front Immunol 13(9): 235.
- Kasprowicz V, A Isa, T Tolfvenstam, K Jeffery, P Bowness, et al. (2006) Tracking of peptide-specific CD4+ T-cell responses after an acute resolving viral infection: a study of parvovirus B19. J Virol 80(22): 11209-11217.
- Shoda H, K Fujio, K Sakurai, K Ishigaki, Y Nagafuchi, et al. (2015) Autoantigen BiP-Derived HLA-DR4 Epitopes Differentially Recognized by Effector and Regulatory T Cells in Rheumatoid Arthritis. Arthritis Rheumatol 67(5): 1171-1181.
- Schulten V, JA Greenbaum, M Hauser, DM McKinney, J Sidney, et al. (2013) Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc Natl Acad Sci U S A 110(9): 3459-3464.
- Rivino L, EA Kumaran, V Jovanovic, K Nadua, EW Teo, et al. (2013) Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol 87(5): 2693-2706.
- Snyder TM, RM Gittelman, M Klinger, DH May, EJ Osborne, et al. (2020) Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv.
- Pianta A, S Arvikar, K Strle, EE Drouin, Q Wang, et al. (2017) Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients With Rheumatoid Arthritis. Arthritis Rheumatol 69(5): 964-975.
- Kamradt T, B Lengl-Janssen, AF Strauss, G Bansal, AC Steere (1996) Dominant recognition of a Borrelia burgdorferi outer surface protein A peptide by T helper cells in patients with treatment-resistant Lyme arthritis. Infect Immun 64(4): 1284-1289.
- Powlson J, D Wright, A Zeltina, M Giza, M Nielsen, et al. (2019) Characterization of Antigenic MHC-Class-I-Restricted T Cell Epitopes in the Glycoprotein of Ebolavirus. Cell Rep 29(9): 2537-2545 e2533.
- Owen DL, SA Mahmud, LE Sjaastad, JB Williams, JA Spanier, et al. (2019) Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol 20(2): 195-205.
- Sette A, J Alexander, J Ruppert, K Snoke, A Franco, et al. (1994) Antigen analogs/MHC complexes as specific T cell receptor antagonists. Annu Rev Immunol 12: 413-431.
- Franco A, C Ferrari, A Sette, FV Chisari (1995) Viral mutations, TCR antagonism and escape from the immune response. Curr Opin Immunol 7(4): 524-531.
-
Seema Rani Padhiary, Sameer Sharma. Role of CRISPR-Cas/9 in Hematological Disorders: A Review. Arch Biomed Eng & Biotechnol. 5(3): 2021. ABEB.MS.ID.000613.
-
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated protein 9 (Cas9) system, Haematological disorders, Gene editing, Haematological malignancies & monogenic,
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






