 Research Article
Research Article
Infrared Spectrum for the New Nanomolecules ASi, CSi, TSi and GSi
Ricardo Gobato1*
Green Land Landscaping and Gardening, Seedling Growth Laboratory, Brazil
Ricardo Gobato, Green Land Landscaping and Gardening, Seedling Growth Laboratory, Bela Vista do Paraíso, 86130-000, Parana, Brazil.
Received Date: April 15, 2020; Published Date: May 21, 2021
Abstract
The core of the work is based on the replacement of carbon atoms by silicon atoms, on the basis of four standard bases of DNA: A, C, G and T (adenine, cytosine, guanine, thymine). Determining with minimum computational methods via ab initio Hartree-Fock methods, infrared spectrum and their peak absorbance frequencies. The option for simple replacement of carbon by silicon is due to the peculiar characteristics between both. Atomic interactions under non-carbon conditions were studied, with only the Hydrogen, Silicon, Nitrogen and Oxygen atoms, in CNTP, for the four standard bases of DNA, A, C, G and T, thus obtaining by quantum chemistry four new compounds, named here as: ASi, CSi, GSi and TSi. Computational calculations admit the possibility of the formation of such molecules, their existence being possible via quantum chemistry. Calculations obtained in the ab initio Unrestricted and Restrict Hartree-Fock method, (UHF and RHF) in the set of bases used Effective core potential (ECP) minimal basis, UHF CEP-31G (ECP split valance) and UHF CEP-121G (ECP triple-split basis), CC-pVTZ (Correlation-consistent valence-only basis sets triple-zeta) and 6-311G**(3df, 3pd) (Gaussian functions quadruple-zeta basis sets).
Keywords:DNA; Adenine; Cytosine; Guanine; Thymine; Hartree-Fock method; Nano-molecule; Infrared spectroscopy; CEP-31G; CEP-121G; CCpVTZ, 6-311G** (3df, 3pd)
Introduction
The Deoxyribonucleic acid (DNA) is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. The A, C, G, and T, - adenine, cytosine, guanine and thymine, respectively, representing the four nucleotide bases of a DNA strand – adenine, cytosine, guanine, thymine – covalently linked to a phosphodiester backbone [1].
The core of the work is based on the replacement of carbon atoms by silicon atoms, on the basis of four standard bases of DNA: A, C, G and T (adenine, cytosine, guanine, thymine). Determining with minimum computational methods via ab initio Hartree-Fock methods, infrared spectrum and their peak absorbance frequencies [2-14].
The option for simple replacement of carbon by silicon [15,16] is due to the peculiar characteristics between both. Atomic interactions under non-carbon conditions were studied, with only the Hydrogen, Silicon, Nitrogen and Oxygen atoms, in STP (Standard Temperature and Pressure), for the four standard bases of DNA, A, C, G and T, thus obtaining by quantum chemistry four new compounds, named here as: ASi, CSi, GSi and TSi. In comparing the carbon and silicon has: the Si lies in the same column of the periodic table of the elements, and it has been investigated as a possible alternative for building up biological molecules in exobiology [17,18]. Silicon based chemistry, however, is by far less flexible than carbon chemistry, not able to form double covalent bonds with the same easiness as C does. Other fact is the larger volume occupied by the external electronic orbitals of silicon tend to reduce the superposition of p orbitals [17,18]. Through the chemical abundances of biolog ical elements in the earth crust, terrestrial life has chosen carbon instead of silicon, in spite of the larger abundance of silicon. This fact suggests that carbon is better suited to form biological molecules [17,18].
However, this paper assumes conditions without the presence of Carbon. Calculations obtained in the ab initio Unrestricted and Restrict Hartree-Fock method, (UHF and RHF). The set of basis used Effective core potential (ECP) minimal basis, UHF CEP-31G (ECP split valance) and UHF CEP-121G (ECP triple-split basis), CCpVTZ (Correlation-consistent valence-only basis sets triple-zeta) and 6-311G**(3df, 3pd) (Gaussian functions quadruple-zeta basis sets) [2-14]. Studies did not reveal any works with characteristics studied here. There is an absence of a referential of the theme, finding only one work in (Gobato et al., 2018-2021) [19-30].
Methods
Hartree-Fock Methods
The molecular Hartree-Fock [2-14] wave function is written as an antisymmetrized product (Slater determinant) of spin-orbitals, each spin-orbital being a product of a spatial orbital ϕi and a spin function (either α or β).
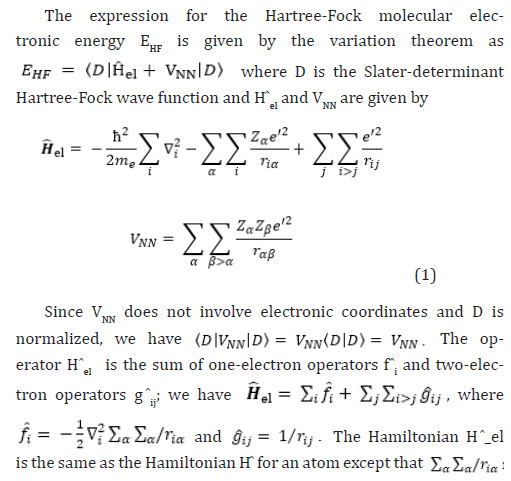
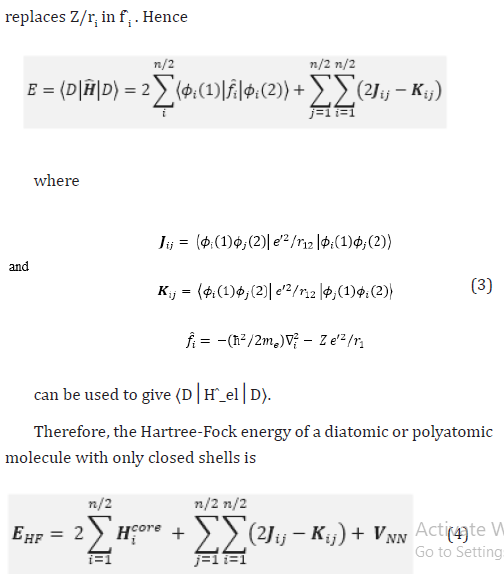
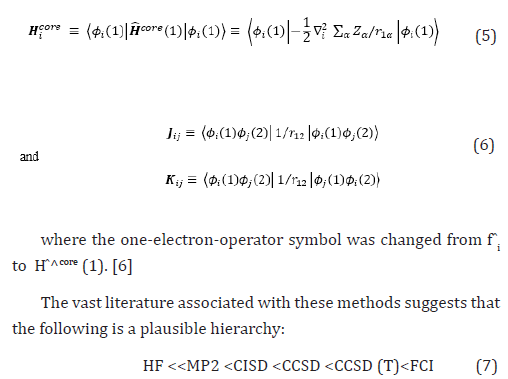
Therefore, the Hartree-Fock energy of a diatomic or polyatomic molecule with only closed shells is
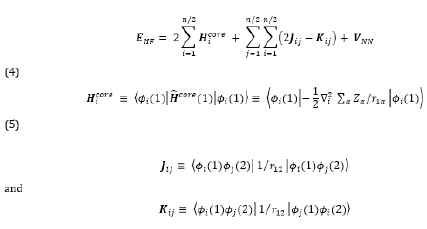
where the one-electron-operator symbol was changed from f ̂i to H ̂core (1) [4].
The vast literature associated with these methods suggests that the following is a plausible hierarchy:
HF < The extremes of ‘best’, FCI, and ‘worst’, HF, are irrefutable, but
the intermediate methods are less clear and depend on the type of
chemical problem being addressed. The use of HF [2-14] in the case
of FCI was due to the computational cost. For calculations the computer used for MD (molecular Dynamics)
was a Desktop with SUSE Linux Enterprise Desktop, AMD Ryzen
7 1800X processor, ASUS Prime A320M-K motherboard, 16GB
of RAM, with 500GB SSD [31]. The ab initio calculations have been performed to study the
equilibrium configuration of ASi, CSi, GSi and TSi molecules. The
set of programs GaussView 5.0.8 [32], Mercury 3.8 [33], Hyper-
Chem 8.0.6 Evaluation [34]. are the advanced semantic chemical
editor, visualization, and analysis platform and GAMESS [15,35] is a
computational chemistry software program and stands for General
Atomic and Molecular Electronic Structure System [15,35], Origin-
Lab 2018 Evaluation [36], BIOVIA Draw 2017 [37], CHARMM22
[38] set of programs were used. The molecular structure of ASi, CSi, GSi and TSi molecules, were
obtained through computationally calculated molecular dynamics,
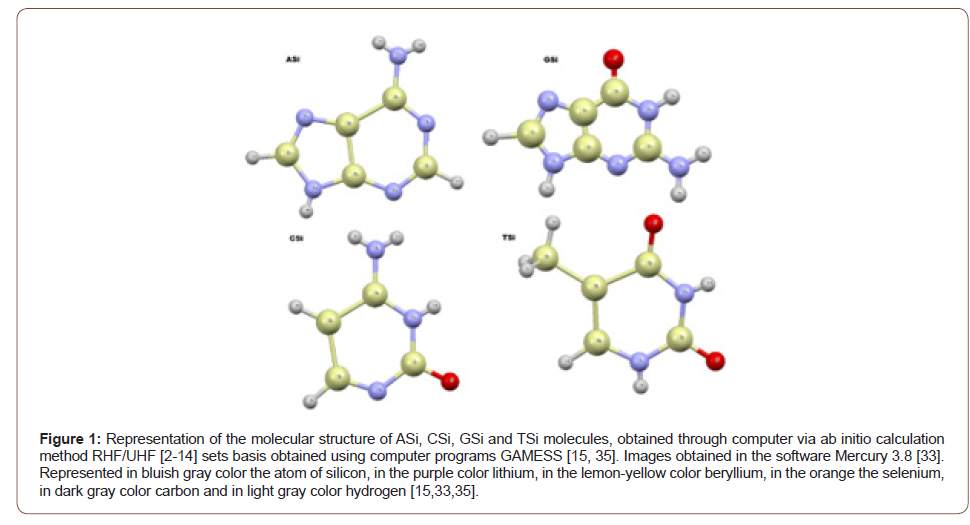
using the ab initio Hartree-Fock (HF) method. The names in Figure (1) of the new molecules obtained ASi, CSi,
GSi and TSi, are: 2,3,4,5,6,7,8,9-octahydro-1H-[1,3,2,4,5] diazatrisilolo[4,5-d]
[1,3,2,4,5,6]diazatetrasilin-8-amine; 2-hydroxy-1,3,2,4,5,6-diazatetrasilinan-4- amine; 8-oxo-3,7-dihydro-[1,3,2,4,5] diazatrisilolo[4,5-d][1,3,2,4,5,6]
diazatetrasilin-6-amine and (2,4-dihydroxy-1,3,2,4,5,6-diazatetrasilinan-5-yl) silane, respectively
(Figure 1). Properties of molecule Asi: • 2,3,4,5,6,7,8,9-octahydro-1H-[1,3,2,4,5] diazatrisilolo [4,5-d]
[1,3,2,4,5,6]diazatetrasilin-8-amine; • PSA(Polar Surface Area): 74.14; • ALogP: 7.1232; • Stereo Center Count: 3; • Hydrogen Acceptor Count: 5; • Hydrogen Donor Count: 5; • Composition: H: 5.9% N: 31.3% Si: 62.8%; • Formula Weight: 223.56421; • Exact Mass: 223.001728116; • Molecular Formula: H13N5Si5 [37].
Properties of molecule CSi: • 2-hydroxy-1,3,2,4,5,6-diazatetrasilinan-4-amine; • PSA: 70.31; • ALogP: 6.022; • Stereo Center Count: 2; • Hydrogen Acceptor Count: 4; • Hydrogen Donor Count: 4; • Composition: H: 6.1% N: 23.2% O: 8.8% Si: 61.9%; • Formula Weight: 181.44883; • Exact Mass: 180.997918122; • Molecular Formula: H11N3OSi4 [37]. Properties of molecule GSi: IUPAC name: • 8-oxo-3,7-dihydro-[1,3,2,4,5] diazatrisilolo[4,5-d][1,3,2,4,5,6]
diazatetrasilin-6-amine; • PSA: 96.16; • ALogP: 5.1999; • Stereo Center Count: 0; • Hydrogen Acceptor Count: 4; • Hydrogen Donor Count: 3; • Composition: H: 2.2% N: 30.3% O: 6.9% Si: 60.7%; • Formula Weight: 231.50009; • Exact Mass: 230.93404248; • Molecular Formula: H5N5OSi5 [37]. Properties of molecule TSi: IUPAC name: • (2,4-dihydroxy-1,3,2,4,5,6-diazatetrasilinan-5-yl) silane; • PSA: 64.52; • ALogP: 8.6476; • Stereo Center Count: 3; • Hydrogen Acceptor Count: 4; • Hydrogen Donor Count: 4; • Composition: H: 5.7% N: 13.2% O: 15.1% Si: 66.1%; • Formula Weight: 212.53497; • Exact Mass: 211.974510294; • Molecular Formula: H12N2O2Si5 [37] (Figure 2-5).
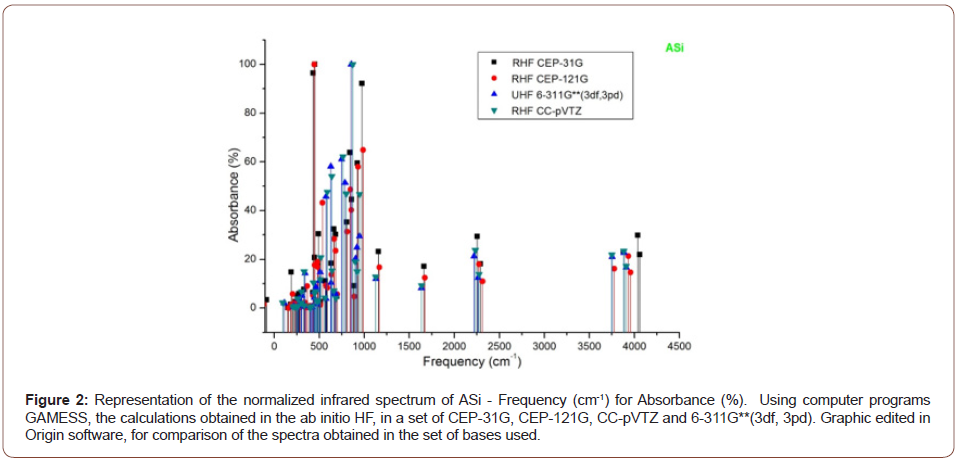
The absorbance peaks for Asi in the set of bases then used between
400 to 100 cm-1, 2250 cm-1 and 3750 to 4100 cm-1, (Figure
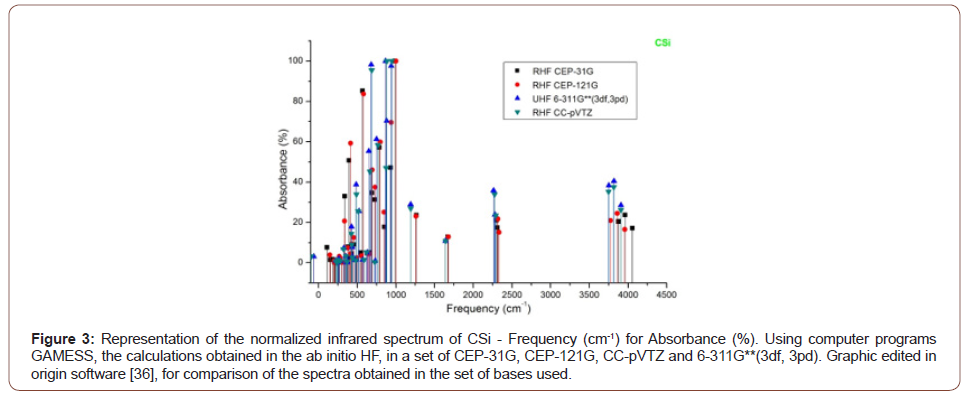
2). The absorbance peaks for Csi in the set of bases then used between
400 to 1000 cm-1, 2250 to 2300 cm-1 and 3750 to 4100 cm-
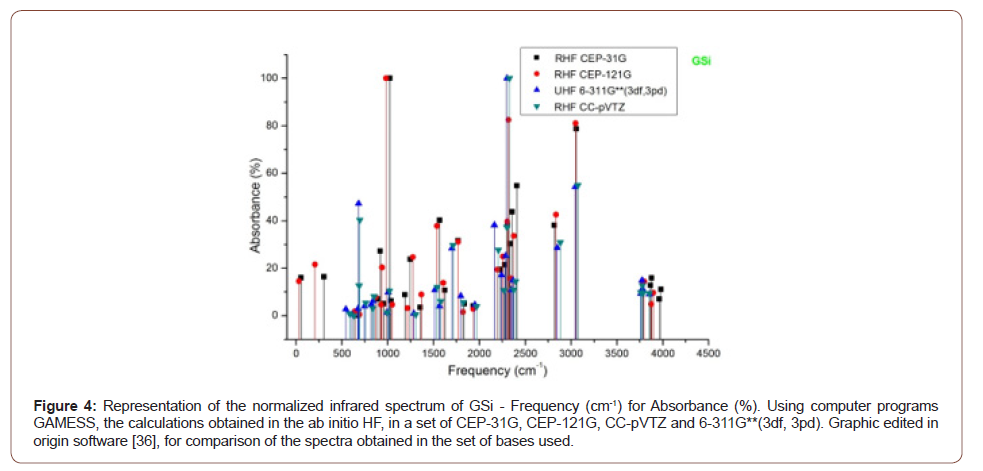
1, (Figure 3). The absorbance peaks for Gsi in the set of bases used then between
the range of 1000 cm-1, 2300 cm-1 and 3000 cm-1, (Figure
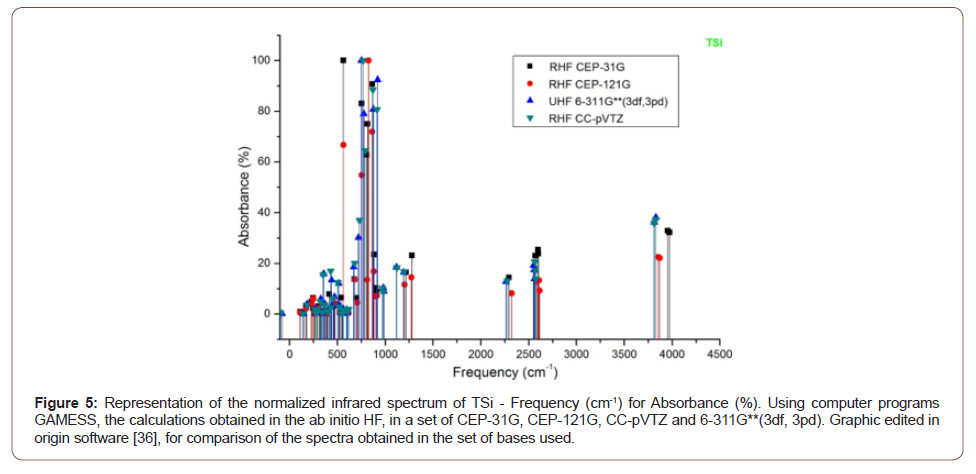
4). The absorbance peaks for Tsi in the set of bases then used between
660 to 950 cm-1, 2250 to 2600 cm-1 and 3500 to 4000 cm-1,
(Figure 5). The fingerprint of the molecules ASi, CSi, GSi and TSI, are
demonstrated in the Tables (1-4) and Figures (2-5). The infrared spectrum was calculated, indicating the characteristic
of the nano-molecule genesis. Characterized its infrared
spectrum, quantically calculated, accepted by quantum chemistry
parameters, with ab initio methods, in the bases CC-pVTZ and
6-311G**(3df, 3pd). An experimental challenge to chemists. Going beyond imagination, now the challenge is to build the
basic structure of is the simulation for building a “new DNA helix”,
based now on the ASi, CSi, GSi and TSi molecules. Limitations our
study has so far been limited to computational simulation via quantum
mechanics e molecular mechanics (QM/MM), an applied theory.
Our results and calculations are compatible and with the theory
of QM/MM, but their physical experimental verification depend on
advanced techniques for their synthesis, obtaining laboratory for
experimental biochemical. None. No conflict of interestHardware and Software
Result
ASi
IUPAC name:
CSi
IUPAC name:
GSi
GSi
TSi
Conclusion and challenges
Acknowledgement
Conflict of Interest
References
- Creative Commons. (CC-BY 4.0). Wikipedia, The Free Encyclopedia, Nucleic acid sequence 2021.
- IN Levine (2003) Quantum Chemistry. Pearson Education (Singapore) Indian Branch, 482 FIE. Patparganj 5th Delhi 110 092, India.
- A Szabo, NS Ostlund (1989) Modern Quantum Chemistry. Dover Publications, New York, USA
- W Kohn, LJ Sham (1965) Self-consistent equations including exchange and correlation effects. Phys Rev 140: A1133.
- JM Thijssen (2001) Computational Physics. Cambridge University Press, Cambridge, USA.
- TH Dunning (1989) Gaussian basis sets for use in correlated molecular calculations, The atoms boron through neon and hydrogen. J Chem Phys (90): 1007-1023.
- DE Woon, TH Dunning (1993) Gaussian-basis sets for use in correlated molecular calculations. The atoms aluminum through argon. J Chem Phys (98): 1358–1371.
- AK Wilson, T van Mourik, TH Dunning (1996) Gaussian basis sets for use in Correlated Molecular Calculations. Sextuple zeta correlation consistent basis sets for boron through neon. J Mol Struct (Theochem), 388: 339-349.
- E Polak (1971) Computational Methods in Optimization. New York 77: 10003,
- T H Dunning, PJ Hay (1977) Modern Theoretical Chemistry. Plenum, New York, 3.
- E Eliav (2013) Elementary introduction to Molecular Mechanics and Dynamics.
- W J Hehre (2003) A Guide to Molecular Mechanics and Quantum Chemical Calculations. Wavefunction Inc Irvine, USA
- MS Gordon, MW Schmidt (2005) Advances in electronic structure theory: GAMESS a decade later. Theory and Applications of Computational Chemistry: the first forty years. In: CE Dykstra, G Frenking, KS Kim GE Scuseria (Eds) 1167-1189.
- F Weigend, R Ahlrichs (2005) Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys Chem Chem Phys (7): 3297–305.
- JJW McDouall (2013) Computational Quantum Chemistry. Molecular Structure and Properties in Silico. The Royal Society of Chemistry, Thomas Graham House, Science Park, Milton Road, Cambridge CB4 0WF, UK.
- S Patai, Z (1989) Rappoport The Chemistry of Organic Silicon Compounds. Series: The Chemistry of functional groups, Wiley.
- AM Shaw (2006) Astrochemistry: From Astronomy to Astrobiology Wiley.
- G Horneck, P Rettberg (2007) Complete Course in Astrobiology. Series: Physics Textbook, Wiley-VCH.
- R Gobato, A Heidari (2017) Calculations Using Quantum Chemistry for Inorganic Molecule Simulation BeLi2 Science Journal of Analytical Chemistry 5(5): 76-85.
- R Gobato, A Heidari (2018) Using the Quantum Chemistry for Genesis of a Nano Biomembrane with a Combination of the Elements Be, Li, Se, Si, C. HJ Nanomed Res 7(4): 241-252.
- R Gobato, A Heidari, A Mitra (2018) The Creation of C13H20BeLi2SeSi. The Proposal of a Bio–Inorganic Molecule, Using Ab Initio Methods for the Genesis of a Nano Membrane. Arc Org Inorg Chem Sci 3(4).
- Ricardo Gobato, Alireza Heidari and Abhijit Mitra. Kurumi: A New Liquid Crystal, (2021).
- R Gobato, MRR Gobato, A Heidari (2018) A Mitra Spectroscopy and Dipole Moment of the Molecule C13H20BeLi2SeSi via Quantum Chemistry Using Ab Initio, Hartree–Fock Method in the Base Set CC–pVTZ and 6–311G**(3df, 3pd). American Journal of Quantum Chemistry and Molecular Spectroscopy, 2(1): 9–17.
- R Gobato, MRR Gobato, A Heidari (2019) Raman Spectroscopy Study of the Nano Molecule C13H20BeLi2SeSi Using ab initio and Hartree–Fock Methods in the Basis Set CC–pVTZ and 6–311G** (3df, 3pd). International Journal of Advanced Engineering and Science 7(1): 14-35,
- R Gobato, MRR Gobato, A Heidari (2019) Attenuated Total Reflection–Fourier Transform Infrared (ATR–FTIR) Spectroscopy Study of the Nano Molecule C13H20BeLi2SeSi Using ab initio and Hartree–Fock Methods in the Basis Set RHF/CC– pVTZ and RHF/6–311G** (3df, 3pd): An Experimental Challenge to Chemists. Chemistry Reports 2(1): 1-26,
- Ricardo Gobato (2019) New Nano-Molecule Kurumi and Raman Spectroscopy using ab initio, Hartree-Fock Method. Am J Biomed Sci & Res 2(4).
- R Gobato, A Heidari (2018) Infrared Spectrum and Sites of Action of Sanguinarine by Molecular Mechanics and ab initio Methods. International Journal of Atmospheric and Oceanic Sciences 2(1): 1-9.
- Gobato, Ricardo, Heidari, Alireza, Mitra Abhijit (2021). Bioinorganic membrane using a new Kurumi liquid crystal.
- A Heidari, R Gobato (2020) Investigation of the internal structure and dynamics of gum cancer cells, tissues and tumors by 13C–NMR spectra of DNA/RNA of gum cancer cells as an essential structural tool for integrative studies of gum cancer cells development. Dent Oral Maxillofac Res 6: 1-3.
- (2021) Creative Commons. (CC-BY 4.0). Wikipedia, The Free Encyclopedia.
- R Dennington, T Keith, J Millam. Gaussview, Version 5, 2009.
- The Cambridge Crystallographic Data Centre (CCDC) (2012) Mercury - crystal structure visualisation, exploration and analysis made easy. Mercury 3.1 Development (Build RC5). The Cambridge Crystallographic Data Centre.Hypercube. Molecular Modeling System. HyperChemTM 7.5 evaluation (2003) Hypercube.
- Origin Lab Ò (2018) Evaluation Licence, Graphing & Analysis, ©OriginLab Corporation,
- MS Gordon (1993) General atomic and molecular electronic structure system (GAMESS). J Comput Chem. 14: 1347-1363.
- BIOVIA Draw 2017 Enterprise. MDL Draw Editor 17.1.0.900. Computational results obtained using software programs from Dassault Systemes BIOVIA. The ab initio calculations were performed with the DMol3 program, and graphical displays generated with Draw. 2017.
- R Brooks, RE Bruccoleri, BD Olafson, DJ States, S Swaminathan, M Karplus (1983) CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations J Comp Chem 4: 187-217.
- IR Brooks, CL Brooks, AD MacKerell, L Nilsson, RJ Petrella, et al. (2009) CHARMM: The Biomolecular Simulation Program, J Comput Chem 30: 1545-1614.
-
Ricardo Gobato. Infrared Spectrum for the New Nanomolecules ASi, CSi, TSi and GSi. Arch Biomed Eng & Biotechnol. 5(3): 2021. ABEB.MS.ID.000614.
-
DNA, Adenine, Cytosine, Guanine, Thymine, Hartree-Fock method, Nano-molecule, Infrared spectroscopy, CEP-31G, CEP-121G, CC-pVTZ, 6-311G** (3df, 3pd)
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






