 Research Article
Research Article
A Novel Computer-Aid Nanotechnology Targeting to Combat Antibiotics Resistance with Superbugs against COVID-19 Pandemic
Hyunjo Kim1* and Jae-Hoon Song2
1School of Pharmacy, Yonsei University, South Korea
2Chairman of Asian-Pacific Research Foundation for Infectious and ANSORP, South Korea
Hyunjo Kim, School of Pharmacy, Yonsei University, South Korea.
Received Date: December 14, 2020; Published Date: January 11, 2021
Abstract
Microbial infections are still among the major public health concerns since several yeasts and fungi, and other pathogenic microorganisms, are responsible for continuous growth of infections and drug resistance against bacteria. During COVID-19 pandemic outbreak antimicrobial resistance rate is fostering the need to develop new strategies against drug-resistant superbugs. Thus, a novel technological approach on improving existing drugs is gaining special interest. Based on nano-medicine performance, successful experiments, and considerable market prospects, nanotechnology will undoubtedly lead a breakthrough in biomedical field also for infectious diseases, as there are several nanotechnological approaches that exhibit important roles in restoring antibiotic activity against resistant bacteria. Hence, the scientific community should also pay attention to developing affordable methodologies so that nanotechnology can reach patients.
Keywords: Nano-antibiotics; Bacterial and virus infections; COVID-19 pandemic; Antimicrobial resistance (AMR); Superbugs; Antimicrobial peptides
Introduction
The current COVID-19 pandemic has placed disproportionate demands on the healthcare infrastructure and economy worldwide, which will negatively impact on the availability of materials as well as the technical capacity for diagnosis, patient care, and treatment of both COVID-19 and non-COVID-19 patients [1-3]. Ubiquitous in nature, antimicrobial resistance (AMR) has existed long before the golden age of antimicrobials [4-8]. Antimicrobial peptides (AMPs) are small peptide-based molecules of 5–100 amino acids in length, with potent and broad-spectrum antimicrobial properties [9,10]. They are part of the innate immune system, which can represent a minimal risk of resistance development. These characteristics contribute to the description of these molecules as promising new molecules in the development of new antimicrobial drugs. However, efforts in developing new medicines have not resulted in any decrease of drug resistance yet. Nanomedicine provides easy access to innovative carriers, which ultimately enable the design and development of targeted delivery systems of the most efficient drugs with increased efficacy and reduced toxicity [11-13]. Furthermore, the increase in AMR has been recognized as a real threat to human and animal health in recent years. It is a problem that has been given the highest priority, uniting nations in the fight against its causes and effects. While antimicrobial agents are beneficial to combat infection, this widespread use contributes to the increase and emergence of novel resistant microbes in virtually all environmental niches. Additionally, the human microbiome is also an important reservoir of AMR with initial exposure occurring in early life. Once seeded with AMR, commensal organisms may be key contributors to the dissemination of resistance due to the interconnectedness of microbial communities. While clinical settings have traditionally been the main focus of the emergence of AMR, non-clinical environments are becoming increasingly recognized as an important factor in the dissemination of antimicrobial resistance genes (ARGs) [14,15]. We are now aware that anthropogenic, commensal, and environmental microorganisms all contribute to the reservoir of ARGs collectively forming the antibiotic resistome [16,17]. The selection pressure driven by the use and misuse of antimicrobials in prescribed human medicine and in disease prevention, control, treatment, and improved growth rates in food-producing animals has significantly contributed to this phenomenon. This has been compounded by the transmission of resistant microbes from person to person and from environmental sources, all influencing the spread of AMR. The microorganisms that inhabit the human body, the human microbiome, and its susceptibility to the spread of AMR, is of particular importance to human health. This increased vulnerability is not only a deadly threat for humans but also constitutes a mobile resistome capable of extending AMR among human populations worldwide. As new resistance mechanisms emerge and spread globally, this will continue to be exacerbated. Classical antibiotic therapy lead to antibiotic resistance and this can prevent by using nano-antibiotics [18-20]. Nano-antibiotics defined as all nano sized materials which have additional antimicrobial activity and can augment the overall efficacy and safety in consumption. We have been observed that using nano encapsulation system can improve efficacy of antibiotics. On the other hand, nano-carriers indicated functional role which can improve drug absorption by enhancing solubility, preventing from drug degradation, controlling intracellular penetration. Therefore, manufacture and development of nano-antibiotics, in addition of prevention of emerging resistant bacteria and reduction the side effects of drugs, can facilitate their storage for long time which can be more economic, although, this science needs investment for developing and becoming commercial technology.
Methods and Result
The source of this study was PubMed and Key words used were Antimicrobial resistant (AMR), Multi-drug resistant (MDR) “Genus name”, Superbugs, Nano-antibiotics, COVID-19.
Detection methods of antimicrobial resistance (AMR)
Our ability to combat the threat of emerging resistance relies on accurate AMR detection methods and the development of therapeutics that function despite the presence of antimicrobial resistance.
High throughput DNA sequencing and bioinformatics have dramatically changed the way AMR is investigated by identifying the genes or mutations that convey resistance [21]. Culture based approaches are used to investigate AMR in the clinical setting using antibiotic sensitivity assays. While this is an effective way to determine if an organism can resist a particular antimicrobial agent, it is dependent on the ability of an organism to be grown in culture, not currently possible for many microbes. The ability to sequence DNA from environmental and clinical samples (meta-genomics and whole genome sequencing) to determine gene content presents the opportunity to identify ARGs as well as understand the mechanism of how organisms acquire ARGs [22]. It also presents tools to determine the extent of spread of ARGs amongst diverse populations without the need to first grow the organism in pure culture.
Antibiotics resistance: dissemination routes of AMR
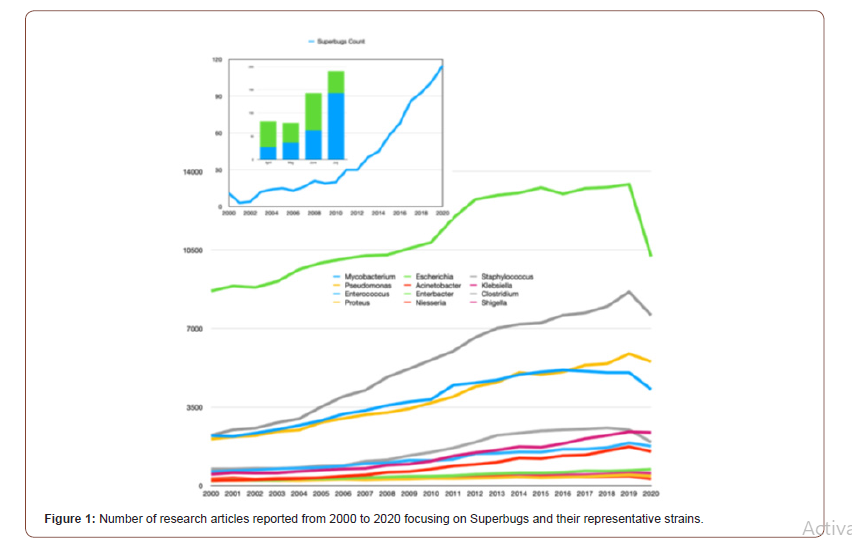
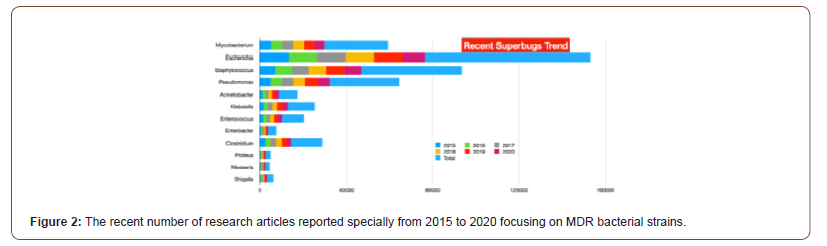
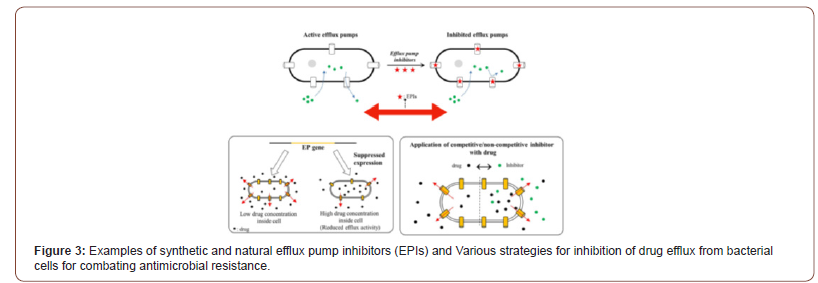
There is no doubt that the COVID-19 pandemic is a clear and present danger in the fight against AMR. As such, it is of the utmost importance that urgent measures are implemented to counteract the risks brought on by the current situation. The commitment of multiple sectors will be crucial, and prominent roles will be played by health institutions, both human and veterinary, and producers of vaccines and antibiotics as well as by governments and international agencies. However, the actions undertaken by individuals will also be paramount in controlling the pandemic and minimizing its impact on our day to day lives. The following months will be decisive in establishing actionable scenarios and preparing clear guidelines that will guarantee that our fight against AMR will continue. Antibiotics, once considered the lifeline for treating bacterial infections, are under threat due to the emergence of threatening antimicrobial resistance (AMR). These drug-resistant microbes (or superbugs as shown in Figure 1) are non-responsive to most of the commonly used antibiotics leaving us with few treatment options and escalating mortality-rates and treatment costs [23-28]. The number of published articles related to superbugs has been remarkably increased for two decades (see Figure1) and the relevant specific strains in the recent trend are presented from 2015 to 2020 specially in Figure 2. The problem is further aggravated by the drying-pipeline of new and potent antibiotics effective particularly against the drug-resistant strains. Multi drug efflux pumps (EPs) are established as principal determinants of AMR, extruding multiple antibiotics out of the cell, mostly in non-specific manner and have therefore emerged as potent drug-targets for combating AMR (see scheme in Figure 3). Collectively, the complex EP assemblies are critical for bacterial pathogenesis, virulence, biofilm formation, and adaptive responses ultimately conferring and defining bacterial AMR [29,30]. EPs are critical for bacterial AMR as they exclude most of the unwanted entities until the cell gets required time for acquiring resistance. Although bacterial AMR has several origins and many adaptive mechanisms are employed by drug-resistant strains against the antibiotics, the intrinsic EPs hold the key (Figure 1-3).
Fighting AMR in the Healthcare Environment
Microbiome-based sanitation approaches and monitoring tools as targeted hygiene to tackle AMR
It was also proposed the hypothesis of further expanding on the importance of exposure to essential microbes in the development of the human immune system. However, with regard to hygiene in the 21st century where humans are still experiencing infectious diseases outbreaks such as SARS [31], MERS [32] and COVID-19 [1,2, 33], newer hypotheses should be proposed rather a more risk assessment approach termed “targeted hygiene (TH)” [34]. Targeted hygiene approach aims to strike a balance between exposure to essential microbes and protection against pathogenic ones and has been used effectively during the previous SARS and Ebolas [35-37] outbreaks approach to further provide insights into AMR implications to human, animal and environment health. On the other hand, healthcare-associated infections (HAIs) affect up to 15% of all hospitalized patients, representing a global concern [38- 40]. Major causes include the persistent microbial contamination of hospital environment, and the growing AMR of HAI-associated microbes [41]. The hospital environment represents in fact a reservoir of potential pathogens, continuously spread by healthcare personnel, visiting persons and hospitalized patients. It showed in Figure 4 that bioburden analyses confirmed TH ability to remodulate the environmental contamination in all settings, as well as to decrease AMR in the contaminant microbial population. In particular, in all treated healthcare structures, a persistent surface contamination was detected, mostly represented by Grampositive bacteria belonging to the Staphylococcus genus, whereas other genera were less represented as shown in Figure 4. Other resistances were however well represented, although to a variable extent, likely depending on the selective pressure exerted by the specific antimicrobial use of each healthcare structure (Figure 4).
The human microbiome as a reservoir for antimicrobial resistance genes
Adopting one health approach at global scale can help in the prevention and control of COVID-19. One important reservoir of AMR is the human microbiome, a complex ecosystem consisting of trillions of microbes closely interacting and exposed to resistance determinants [42]. It is estimated that the human microbiome harbors about 3.3 million non-redundant genes, which represents a gene set 150 times larger than that of the human host. This extensive genetic diversity, along with the 10– 100 trillion microbial cells making up this micro-ecosystem, extend the host genetic, metabolic and immune capabilities primarily through regulating energy metabolism and helping to shape innate and adaptive immunity [43]. The confined locations of this high cell and genetic density also provide ideal conditions for genetic exchange between transient and residential microbes, and among residential microbes. For instance, the human distal gut harbors about 1014 microbial cells representing around 400 different bacterial phylotypes [44]. While the GI microbiome harbors commensal organisms, it also contains undesirable genetic traits linked to microbial virulence and AMR [45]. Figure 5 (top) shows the distribution of nucleotide identities for 93 resistance genes identified from DNA extracted directly from saliva and fecal samples and 114 resistance genes identified from cultured aerobic gut isolates to the most similar resistance gene harbored by a pathogenic isolate in NCBI GenBank. Overlap of resistance genes identified from the human microbiomes of two healthy unrelated individuals. resistance genes from two individuals are compared to each other and defined as being shared if they have greater than 90% sequence identity at the nucleotide level [46]. These resistance genes identified using meta-genomic functional selections are shared between two individuals to a much lesser extent than resistance genes identified from cultured gut isolates (see Figure 5 bottom). Notably, all resistance genes that are shared between the two individuals also have high sequence identity (>90% at the nucleotide level) to previously characterized genes in NCBI GenBank (Figure 5).
Interfering with the stability of the mobilome for antibiotic resistance
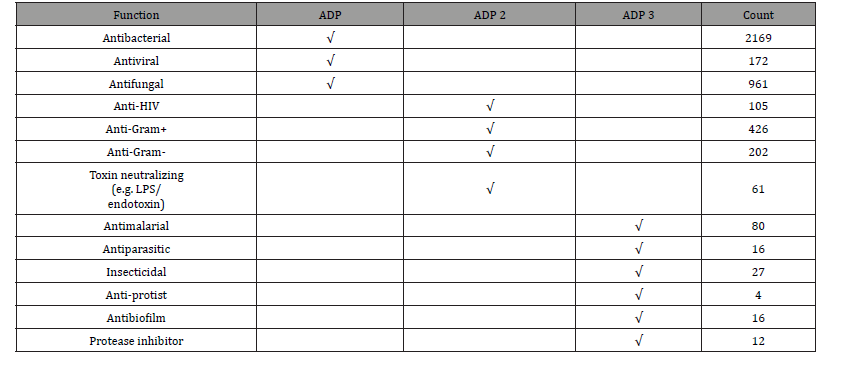
In this context, the collective genetic arsenal responsible for conferring antibiotic resistance (AR) through inactivation and/or removal of antibiotics is commonly referred to as the resistome. The occurrence of AR genes may increase the ecological fitness of a bacterium and thus its ability to colonize and persist in a specific environment. Putative AR genes encoding transporters can be excluded from this analysis due to the inaccuracy of their bioinformatic prediction. Thus, spread of AMR is largely due to genetic exchange mediated by the mobilome since resistance genes are present in plasmids, phages, integrative and mobilizable elements (IMEs), and conjugative elements (ICEs) [47,48]. Thus, understanding the mobilome and how it moves among bacteria is essential to deal with AMR. However, in addition to being acquired, mobile genetic elements (MGEs) can also be lost, and this may constitute a yet unexplored way to cope with AMR by interfering with the mechanisms that ensure the conservation of the mobilome (see Figure 6). It was predicted resistome of the Bifidobacterium genus and abundance of different predicted antibiotic resistance gene classes identified among the 625 analyzed Bifidobacterium genomes. In addition to it, these illustrations in Figure 5 display unique mobile genetic hotspots identified in B. longum subsp. longum E18 and B. parmae LMG 30,295 strains. In particular, inducing plasmid loss could be a strategy worth exploration. We can conceive this approach as follows: instead of interfering with plasmid conjugation that lasts from minutes to a few hours, or with the outcome of mutations that may occur within minutes, it could be worthwhile to interfere with plasmid-chromosome crosstalk that lasts a lifetime, as long as the plasmid is not lost [49]. Then, the identification and employment of new molecules able to interfere with plasmid stability, alone or in combination with conjugation inhibitors (COINs), could lead to a substantial impulse in the battle against the spread of AMR [50] (described in Figure 7). Table 1 summarized the possibility of new molecules entity as antimicrobial peptide against AMR using computer aid structure database construction [51-63] (Figure 6,7) (Table 1,2).
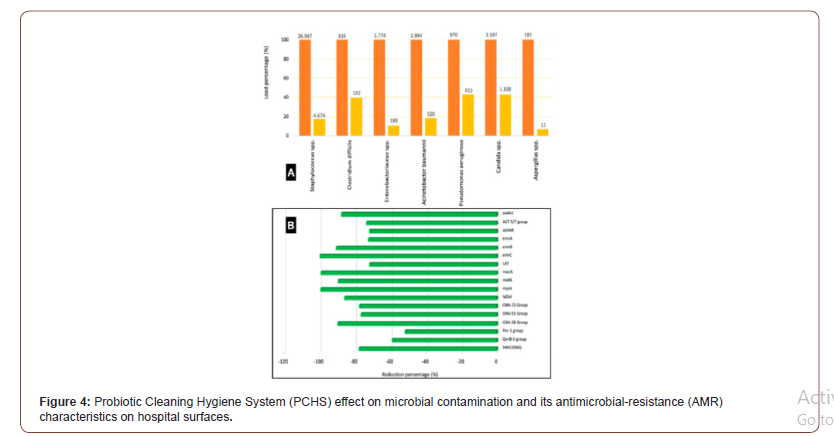
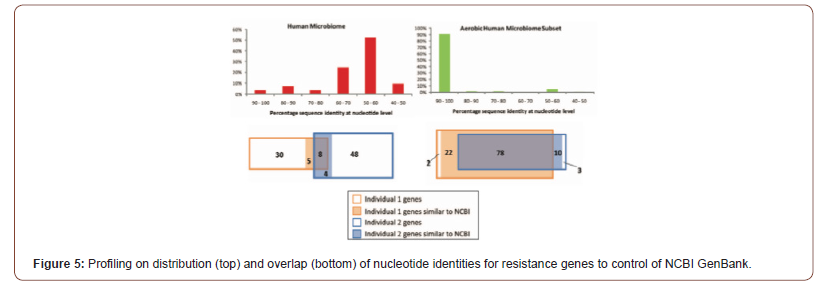
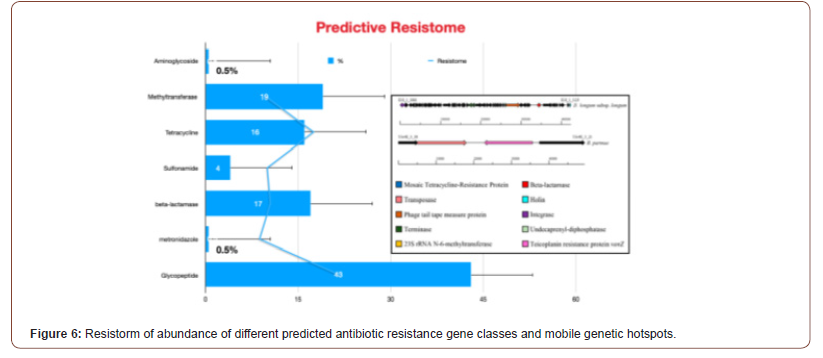
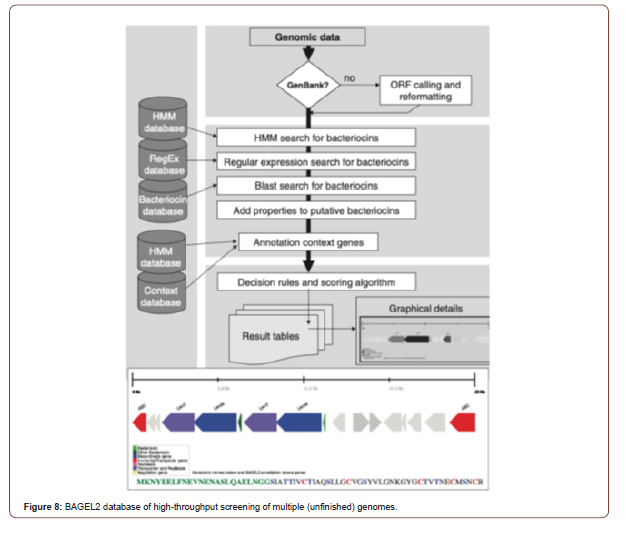
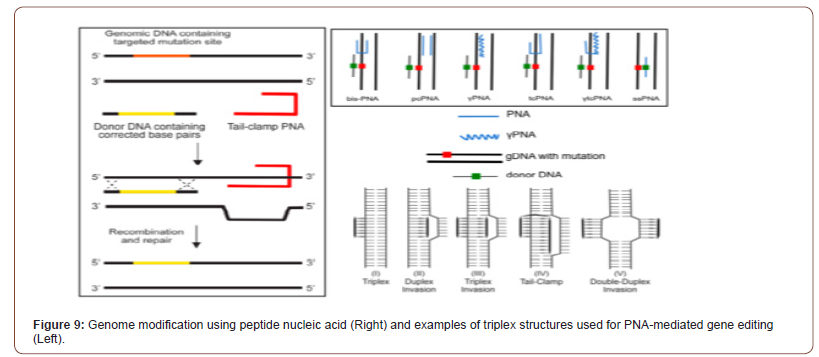
Table 1: Characteristics and description of currently available AMPs databases.
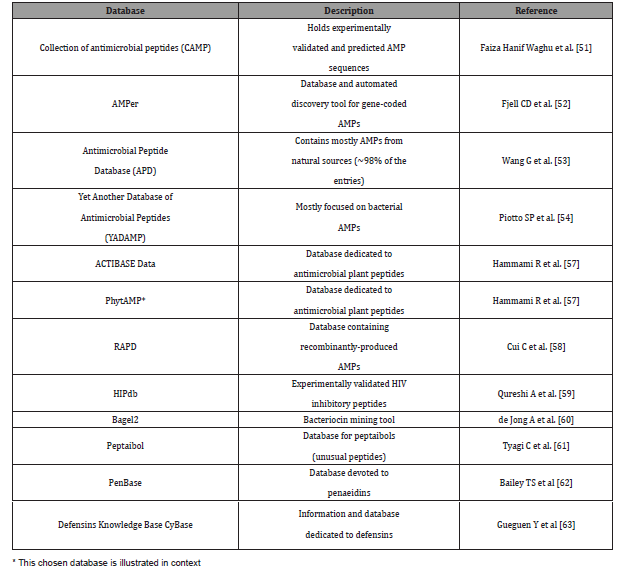
Table 2: Expansion of searchable peptide functions in the antimicrobial peptide database.
Miscellaneous Strategies
A Novel targeting virulence trait
Additional approaches which may be complementary to the use of antibiotics envisage interfering with virulence traits rather than with biosynthetic or growth pathways. In this approach, the aim is not to kill the pathogenic bacteria, but rather to diminish their virulence, thus avoiding selection of resistances and allowing the combination of these strategies with the use of antibiotics. Alternatively, targeting genes involved in bacterial motility (e.g., flagella, fimbriae) or hindering the assembly of the proteins participating in these organelles could lead to a substantial reduction in the dissemination of the infection (see Figure 8 including graphical representation). The input genome data can be a single or multi-entry GenBank file, in case of non-annotated data or, if re-annotation is desired, an ORF calling can be performed via the BAGEL2 [60] web site which is graphical output for putative bacteriocin and was identified through the new MA-2PEPA motif [64]. Amino acids in the leader sequence of the putative bacteriocin are indicated in green. Amino acids potentially involved in lanthionine ring formation are marked in red (cysteine) and blue (serine and threonine). The annotation of putative bacteriocins and their genomic context genes followed by calculation of score for each candidate and generation of detailed reports (Figure 8).
Antisense RNAs (asRNAs)
Alternatives to the above strategies are under intense research, especially the employment of asRNAs to shut down translation. In this field, the more efficient approaches so far include the use of modified asRNAs coupled to oligodeoxynucleotides to silence essential genes at mRNA: DNA levels. However, these DNA: RNA hybrid molecules are easily degraded by intracellular nucleases (see Figure 9). To avoid this inconvenience, variants of these molecules have been sought, especially molecules that are formed by peptide-nucleic acids (PNAs) [65-67]. The PNAs are analogues of nucleic acids in which the ribose/deoxyribose-phosphate backbone has been replaced by a flexible peptide polymer to which the bases are attached. Many multi-drug resistant bacteria have been increased and designing novel components is necessary for confrontation these bacteria. For PNA targeting gene, peptide- PNAs (PPNA) activity has been affected by sequence-based specific target and dose-dependent-based manner. Applications of these components are wide while, and one of the important applications of these components is inhibition of gene expression and knock downing the target gene follow as inhibition of bacterial growth. Some novel opportunities for advancement and the design ultranarrow- spectrum antimicrobial drugs against multi-drug can be accessible by utilizing PNA against necessary genes of pathogens. This study highlights the novel nanoparticle-based delivery strategies employed for delivery of regular as well as chemically modified PNAs [68] (Figure 9).
Nano-Antibiotics: Nano carriers for COVID-19 Control
Bacteria show resistance to antibiotics drugs through a variety of mechanisms. Moreover, the development of even new mechanisms of resistance has resulted in the simultaneous development of resistance to several antibiotic classes creating very dangerous multidrug-resistant (MDR) bacterial strains. However, when bacteria are drug-resistant it does not mean that they stop responding to an antibiotic, but that occurs only at higher concentrations. Of greater concern are cases of acquired resistance, where initially susceptible populations of bacteria become resistant to an antibacterial agent, in particular antibiotics, and proliferate and spread under the selective pressure of use of that drug. One approach to address this challenge is to design analogues of drugs that of use of that drug. One approach to address this challenge is to design analogues of drugs are already in clinical use and that have activity against resistant organisms. However, bacteria are constantly succeeding to develop a resistant mechanism to new antibiotic drugs as well as to their analogues. Nanotechnology offers opportunities to re-explore the biological properties of already known antimicrobial compounds such as antibiotics by manipulating their size to modify their effect [69,70]. antimicrobial compounds such as antibiotics by manipulating their size to modify their effect. The combination of AMPs with nanomaterials is not new.
Drug Delivery System for Nano-Antibiotics
Nanomaterials, which either show antimicrobial activity by themselves or elevate the effectiveness and safety of antibiotics administration, are called “nano-antibiotics” [71,72] and their capability of controlling infections in vitro and in vivo has been explored and demonstrated. Unlike many antimicrobial agents currently being used in the clinic, antimicrobial NPs may not pose direct and acute adverse effects, although potential toxicity upon long-term exposure is questionable. Most importantly, antimicrobial NPs tackle multiple biological pathways found in broad species of microbes importantly, many concurrent mutations would have to occur to develop resistance against NPs’ activities. Preparation of antimicrobial NPs could be cost-effective, compared to antibiotics synthesis, antimicrobial activities. compared to furthermore they are stable enough for long-term storage with a prolonged shelf-life. The possible nano-nano-carriers are illustrated in Figure 10. In addition, some NPs can withstand harsh conditions, such as hightemperature conventional antibiotics are inactivated. Antibiotics delivery using nanomaterials offer multiple sterilization, under which conventional antibiotics are inactivated. Consequently, technological approach on improving existing drugs is gaining special interest. Nanomedicine provides easy access to innovative carriers, which ultimately enable the design and development of targeted delivery systems of the most efficient drugs with increased efficacy and reduced toxicity. Nanotechnology will undoubtedly lead a breakthrough in biomedical field also for infectious diseases, as there are several nanotechnological approaches that exhibit important roles in restoring antibiotic activity against resistant bacteria [73] (Figure 10).
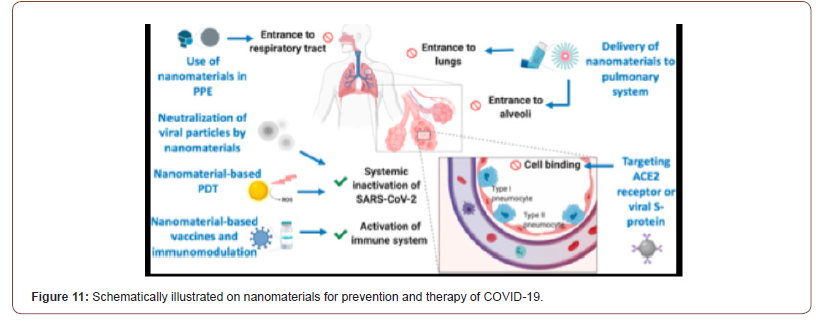
Nanotechnology-enveloped approaches against the COVID-19 Pandemic
Nanotechnology offers a number of solutions to fight viruses, both outside and inside the host, and several nanotechnologiesbased platforms [74-76] have already been successful in preclinical studies to counter several human viral pathogens such as HIV [77], human papilloma virus [78], herpes simplex [79,80], and respiratory viruses [80]. Nanotechnology-based approaches should be leveraged to help the fight against COVID-19 as well as any future pandemics, in a number of ways. Nanomaterials for prevention and therapy of COVID-19. Integrating nanomaterials into personal protective equipment (PPE) can prevent the entrance of SARS-CoV-2 in the respiratory system. Nanomaterials could also be used to deliver drugs to the pulmonary system via inhalators. Cellular binding of viral particles at the alveoli can be inhibited using targeted nanoparticles (NPs) against angiotensin-converting enzyme 2 (ACE2) receptors or viral S protein. Various mechanisms can be used to inactivate viral particles systemically such as using neutralizing NPs or photocatalytic nanomaterials. Nanomaterialbased vaccines or immunomodulation can be used to prevent SARS-CoV-2 infection or even to boost the immune response during infection. PDT, photodynamic therapy (see Figure 11). This crisis has also highlighted the importance of rapid prototyping/ manufacturing for addressing unforeseen needs, such as in case of a pandemic, where large-scale production of equipment including ventilators and personal protective equip- ment (PPE) is urgently needed and nanotechnology may aid (e.g., in providing readily synthesizable materials for equipment manufacturing as well as for the improvement of their efficiency and durability). In the next sections, we expand and elaborate on the specific contributions that nanotechnology can offer to counter COVID-19 and similar pandemics (Figure 11).
Discussion
Antimicrobial resistance is a complex problem with many diverse contributing factors. It is major cause of health concerns adding cost to oneself and to the community, directly or indirectly. When acquired by pathogens however, AMR becomes a serious public health threat worldwide. Antimicrobial resistance (AMR) is a global health concern of increasing magnitude, specially in COVID-19 pandemic era. Prevention is still the best tool to reduce the infection spread and thereby AMR. Along with rational use of existing antimicrobial drugs, development of new effective compounds and new diagnostic technology is the need. Joint efforts from patients, prescribers and individuals to international regulators and policy makers are needed to fight against the globally spreading antimicrobial resistance.
Conclusion
Consequently, antimicrobial agents have been used for decades to treat infectious disease, which has successfully reduced illness and death from infectious microbial species. It has become well recognized that microorganisms including bacteria, fungi, parasites, and viruses are capable of developing resistance that make these agents less effective. While resistance mechanisms are emerging in all groups of microbes, an important root cause contributing to this increase is the global over- and misuse of antibiotics. As such, this research focuses on the mechanisms that define the development, transfer, and subsequent detection of bacterial resistance in the human microbiome with an emphasis on the impact of AMR in human health. The emergence of AMR is superseding our ability to combat infection and the development of novel antibiotic therapies such as nano antibiotics and peptide-nucleic acids is essential to address this growing threat.
Acknowledgement
None.
Conflicts of Interest
None to declare.
References
- Driggin E, Madhavan MV, Bikdeli B, Taylor Chuich, Justin Laracy et al. (2020) Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the COVID-19 Pandemic. J Am Coll Cardiol 75(18): 2352-2371.
- Zhao D, Yao F, Wang L, Ling Zheng, Yongjun Gao, et al. (2020) A Comparative Study on the Clinical Features of Coronavirus 2019 (COVID-19) Pneumonia with Other Pneumonias. Clin Infect Dis 71(15): 756-761.
- Helms J, Tacquard C, Severac F, Ian Leonard-Lorant, Mickael Ohana, et al. (2020) High risk of thrombosis in patients with severe SARSCoV-2 infection: a multicenter prospective cohort study. Intensive Care Med 46(6): 1089-1098.
- Clancy CJ, Nguyen MH (2020) COVID-19, superinfections and antimicrobial development: What can we expect? Clin Infect Dis 524.
- Nieuwlaat R, Mbuagbaw L, Mertz D, Lori Burrows, Dawn ME Bowdish, et al. (2020) COVID-19 and Antimicrobial Resistance: Parallel and Interacting Health Emergencies. Clin Infect Dis 773.
- Rawson TM, Ming D, Ahmad R, Moore LSP, Holmes AH (2020) Antimicrobial use, drug-resistant infections and COVID-19. Nat Rev Microbiol 18(8): 409-410.
- Bengoechea JA, Bamford CG (2020) SARS-CoV-2, bacterial co-infections, and AMR: the deadly trio in COVID-19? EMBO Mol Med 12(7): e12560.
- Rodriguez-Alvarez M, Lopez-Vidal Y, Soto-Hernandez JL, Miranda-Novales MG, FloresMoreno K, et al. (2020) COVID-19: Clouds Over the Antimicrobial Resistance Landscape. Arch Med Res S0188-4409(20): 31128-0.
- Elnagdy S, AlKhazindar M (2020) The Potential of Antimicrobial Peptides as an Antiviral Therapy against COVID-19. ACS Pharmacol Transl Sci 3(4): 780-782.
- Maiti BK (2020) Potential Role of Peptide-Based Antiviral Therapy Against SARS-CoV-2 Infection. ACS Pharmacol Transl Sci 3(4): 783-785.
- Hu TY, Frieman M, Wolfram J (2020) Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat Nanotechnol 15(4): 247-249.
- Abd Ellah NH, Gad SF, Muhammad K, E Batiha G, Hetta HF (2020) Nanomedicine as a promising approach for diagnosis, treatment and prophylaxis against COVID-19. Nanomedicine (Lond) 15(21): 2085-2102.
- Weiss C, Carriere M, Fusco L, Ilaria Capua, Jose Angel Regla-Nava et al. (2020) Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 14(6): 6383-6406.
- Ramadan HK, Mahmoud MA, Aburahma MZ, Amal A Elkhawaga, Mohamed A El-Mokhtar, et al. (2020) Predictors of Severity and Co-Infection Resistance Profile in COVID-19 Patients: First Report from Upper Egypt. Infect Drug Resist 13: 3409-3422.
- Tola M, Ajibola O, Idowu ET, Omidiji O, Awolola ST, et al. (2020) Molecular detection of drug resistant polymorphisms in Plasmodium falciparum isolates from Southwest, Nigeria. BMC Res Notes 13(1): 497.
- Crofts TS, Gasparrini AJ, Dantas G (2017) Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol 15(7): 422-434.
- Gasparrini AJ, Crofts TS, Gibson MK, Tarr PI, Warner BB, et al. (2016) Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes 7(5): 443-449.
- Dua K, Gupta G, Koteswara Rao N, Bebawy M (2018) Nano-antibiotics: a novel approach in treating P. aeruginosa biofilm infections. Minerva Med 109(5): 400.
- Hadinoto K, Cheow WS (2014) Nano-antibiotics in chronic lung infection therapy against Pseudomonas aeruginosa. Colloids Surf B Biointerfaces 116: 772-785.
- Jamil B, Bokhari H, Imran M (2017) Mechanism of Action: How Nano-Antimicrobials Act? Curr Drug Targets 18(3): 363-373.
- Pasquali F, Do Valle I, Palma F (2019) Application of different DNA extraction procedures, library preparation protocols and sequencing platforms: impact on sequencing results. Heliyon 5(10): e02745.
- Shaw PJ, Chaotheing S, Kaewprommal P, Jittima Piriyapongsa, Chayaphat Wongsombat et al. (2015) Plasmodium parasites mount an arrest response to dihydroartemisinin, as revealed by whole transcriptome shotgun sequencing (RNA-seq) and microarray study. BMC Genomics 16: 830.
- Rudramurthy GR, Swamy MK, Sinniah UR, Ghasemzadeh A (2016) Nanoparticles: Alternatives Against Drug-Resistant Pathogenic Microbes. Molecules 21(7): 836.
- Saadeh HA, Mubarak MS (2017) Hybrid Drugs as Potential Combatants Against Drug-Resistant Microbes: A Review. Curr Top Med Chem 17(8): 895-906.
- Honigsbaum M (2018) Superbugs and us. Lancet 391(10119): 420.
- Bravo A, Ruiz-Cruz S, Alkorta I, Espinosa M (2018) When Humans Met Superbugs: Strategies to Tackle Bacterial Resistances to Antibiotics. Biomol Concepts 9(1): 216-226.
- Hamad M, Al-Marzooq F, Orive G, Al-Tel TH (2019) Superbugs but no drugs: steps in averting a postantibiotic era. Drug Discov Today 24(12): 2225-2228.
- Khan SN, Khan AU (2016) Breaking the Spell: Combating Multidrug Resistant 'Superbugs'. Front Microbiol 7: 174.
- Kardan-Yamchi J, Kazemian H, Haeili M, Harati AA, Amini S, et al. (2019) Expression analysis of 10 efflux pump genes in multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis clinical isolates. J Glob Antimicrob Resist 17: 201-208.
- Palazzotti D, Bissaro M, Bolcato G, Andrea Astolfi 2, Tommaso Felicetti, et al. (2019) Deciphering the Molecular Recognition Mechanism of Multidrug Resistance Staphylococcus aureus NorA Efflux Pump Using a Supervised Molecular Dynamics Approach. Int J Mol Sci 20(16): 4041.
- Satija N, Lal SK (2007) The molecular biology of SARS coronavirus. Ann N Y Acad Sci 1102(1): 26-38.
- Mackay IM, Arden KE (2015) MERS coronavirus: diagnostics, epidemiology and transmission. Virol J 12: 222.
- Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K (2020) COVID-19 (Novel Coronavirus 2019) - recent trends. Eur Rev Med Pharmacol Sci 24(4): 2006-2011.
- The case for Targeted Hygiene. Perspect Public Health 139(5): 219-221.
- Jadav SS, Kumar A, Ahsan MJ, Jayaprakash V (2015) Ebola virus: current and future perspectives. Infect Disord Drug Targets 15(1): 20-31.
- Baseler L, Chertow DS, Johnson KM, Feldmann H, Morens DM (2017) The Pathogenesis of Ebola Virus Disease. Annu Rev Pathol 12: 387-418.
- Takada A (2015) Uirusu. 65(1): 61-70.
- Al-Tawfiq JA, Tambyah PA (2014) Healthcare associated infections (HAI) perspectives. J Infect Public Health 7(4): 339-344.
- Weiner LM, Webb AK, Limbago B, Margaret A Dudeck, Jean Patel, et al. (2016) Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol 37(11): 1288-1301.
- Weber DJ, Anderson D, Rutala WA (2013) The role of the surface environment in healthcare-associated infections. Curr Opin Infect Dis 26(4): 338-344.
- D Accolti M, Soffritti I, Mazzacane S, Caselli E (2019) Fighting AMR in the Healthcare Environment: Microbiome-Based Sanitation Approaches and Monitoring Tools. Int J Mol Sci 20(7): 1535.
- Brinkac L, Voorhies A, Gomez A, Nelson KE (2017) The Threat of Antimicrobial Resistance on the Human Microbiome. Microb Ecol 74(4): 1001-1008.
- Sharma A, Gilbert JA (2018) Microbial exposure and human health. Curr Opin Microbiol 44: 7987.
- Rivett DW, Bell T (2018) Abundance determines the functional role of bacterial phylotypes in complex communities. Nat Microbiol 3(7): 767-772.
- Taft DH, Liu J, Maldonado-Gomez MX, Samir Akre, M Nazmul Huda, et al. (2018) Bifidobacterial Dominance of the Gut in Early Life and Acquisition of Antimicrobial Resistance. mSphere 3(5): e00441-18.
- Kunishima H, Ishibashi N, Wada K, Kentaro Oka, Motomichi Takahashi, et al. (2019) The effect of gut microbiota and probiotic organisms on the properties of extended spectrum beta-lactamase producing and carbapenem resistant Enterobacteriaceae including growth, beta-lactamase activity and gene transmissibility. J Infect Chemother 25(11): 894-900.
- Huang J, Ma J, Shang K, et al. (2016) Evolution and Diversity of the Antimicrobial Resistance Associated Mobilome in Streptococcus suis: A Probable Mobile Genetic Elements Reservoir for Other Streptococci. Front Cell Infect Microbiol 6: 118.
- Slizovskiy IB, Mukherjee K, Dean CJ, Boucher C, Noyes NR (2020) Mobilization of Antibiotic Resistance: Are Current Approaches for Colocalizing Resistomes and Mobilomes Useful? Front Microbiol 11:1376.
- Johansson MHK, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, et al. (2020) Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: Mobile Element Finder. J Antimicrob Chemother 390.
- Hammami R, Ben Hamida J, Vergoten G, Fliss I (2009) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 37(Database issue): D963-D968.
- Waghu FH, Idicula-Thomas S (2020) Collection of antimicrobial peptides database and its derivatives: Applications and beyond. Protein Sci 29(1): 36-42.
- Fjell CD, Hancock RE, Cherkasov A (2007) AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics 23(9): 1148-1155.
- Wang G, Li X, Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44(D1): D1087-D1093.
- Piotto SP, Sessa L, Concilio S, Iannelli P (2012) YADAMP: yet another database of antimicrobial peptides. Int J Antimicrob Agents 39(4): 346-351.
- Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I (2010) BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 10: 22.
- Hammami R, Zouhir A, Ben Hamida J, Fliss I (2007) BACTIBASE: a new web-accessible database for bacteriocin characterization. BMC Microbiol 7: 89.
- Hammami R, Ben Hamida J, Vergoten G, Fliss I (2009) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res 37(Database issue): D963-D968.
- Cui C, Li Y, Liu Y, Xiaojie Li, Shiju Luo, et al. (2017) Determination of genetic diversity among Saccharin-a germplasm using ISSR and RAPD markers. C R Biol 340(2): 76-86.
- Qureshi A, Thakur N, Kumar M (2013) HIPdb: a database of experimentally validated HIV inhibiting peptides. PLoS One 8(1): e54908.
- De Jong A, Van Heel AJ, Kok J, Kuipers OP (2010) BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res 38(Web Server issue): W647-W651.
- Tyagi C, Marik T, Vagvolgyi C, Kredics L, Otvos F (2019) Accelerated Molecular Dynamics Applied to the Peptaibol Folding Problem. Int J Mol Sci 20(17): 4268.
- Bailey TS, Stone JY (2017) A novel pen-based Bluetooth-enabled insulin delivery system with insulin dose tracking and advice. Expert Opin Drug Deliv 14(5): 697-703.
- Gueguen Y, Bernard R, Julie F, Schmitt Paulina, Destoumieux-Garzon Delphine, et al. (2009) Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol Immunol 46(4): 516-522.
- Van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP (2013) BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41(Web Server issue): W448-W453.
- Narenji H, Gholizadeh P, Aghazadeh M, Rezaee MA, Asgharzadeh M, et al. (2017) Peptide nucleic acids (PNAs): currently potential bactericidal agents. Biomed Pharmacother. 93: 580-588.
- Ricciardi AS, Quijano E, Putman R, Saltzman WM, Glazer PM (2018) Peptide Nucleic Acids as a Tool for Site-Specific Gene Editing. Molecules 23(3): 632.
- Lee HT, Kim SK, Yoon JW (2019) Antisense peptide nucleic acids as a potential anti-infective agent. J Microbiol. 57(6): 423-430.
- Gupta A, Bahal R, Gupta M, Glazer PM, Saltzman WM (2016) Nanotechnology for delivery of peptide nucleic acids (PNAs). J Control Release 240: 302-311.
- Zhang Y, Yuan Y, Chen W, Fan J, Lv H, et al. (2019) Integrated nanotechnology of synergism-sterilization and removing-residues for neomycin through nano-Cu2O. Colloids Surf B Biointerfaces 183: 110371.
- Ojemaye MO, Adefisoye MA, Okoh AI (2020) Nanotechnology as a viable alternative for the removal of antimicrobial resistance determinants from discharged municipal effluents and associated watersheds: A review. J Environ Manage. 275: 111234.
- Jamil B, Bokhari H, Imran M (2017) Mechanism of Action: How Nano-Antimicrobials Act? Curr Drug Targets 18(3): 363-373.
- Dua K, Gupta G, Koteswara Rao N, Bebawy M (2018) Nano-antibiotics: a novel approach in treating P. aeruginosa biofilm infections. Minerva Med 109(5): 400.
- Zhang L, Wang Y, Wang C, Ming He, Jiangshan Wan, et al. (2020) Light-Activable On-Demand Release of Nano-Antibiotic Platforms for Precise Synergy of Thermochemotherapy on Periodontitis. ACS Appl Mater Interfaces 12(3): 3354-3362.
- Yadavalli T, Shukla D (2017) Role of Metal and Metal Oxide Nanoparticles as Diagnostic and Therapeutic Tools for Highly Prevalent Viral Infections. Nanomedicine 13(1): 219-230.
- Singh L, Kruger HG, Maguire GEM, Govender T, Parboosing R (2017) The Role of Nanotechnology in the Treatment of Viral Infections. Ther Adv Infect Dis 4: 105-131.
- Jackman JA, Lee J, Cho NJ (2016) Nanomedicine for Infectious Disease Applications: Innovation towards Broad-Spectrum Treatment of Viral Infections. Small 12(9): 1133-1139.
- Adesina SK, Akala EO (2015) Nanotechnology Approaches for the Delivery of Exogenous siRNA for HIV Therapy. Mol Pharmaceutics 12 (12): 4175-4187.
- Chen J, Gu W, Yang L, Renfu Shao, Kewei Xu, et al. (2015) Nanotechnology in the management of cervical cancer. Rev Med Virol 25 Suppl 1: 72-83.
- Kondel R, Shafiq N, Kaur IP, Mini P Singh, Avaneesh K Pandey, et al. (2019) Effect of Acyclovir Solid Lipid Nanoparticles for the Treatment of Herpes Simplex Virus (HSV) Infection in an Animal Model of HSV-1 Infection. Pharm Nanotechnol 7(5): 389-403.
- Al-Halifa S, Gauthier L, Arpin D, Bourgault S, Archambault D (2019) Nanoparticle-Based Vaccines Against Respiratory Viruses. Front Immunol 10: 22.
-
Hyunjo Kim, Jae-Hoon Song. A Novel Computer-Aid Nanotechnology Targeting to Combat Antibiotics Resistance with Superbugs against COVID-19 Pandemic. 5(1): 2021. ABEB.MS.ID.000604.
-
Nano-antibiotics, Bacterial and virus infections, COVID-19 pandemic, Antimicrobial resistance (AMR), Superbugs, Antimicrobial peptides, Nanomaterial, Receptor, Pathogens, Antibacterial agent
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






