 Research Article
Research Article
1H NMR Spectroscopy of the New Xalapa Molecule
Ricardo Gobato1*, Alireza Heidari2, Lauro Figueroa Valverde3, Abhijit Mitra4, Marcela Rosas Nexticapa5, Maria Lopez Ramos3, Maria Virginia del Socorro Mateu Armad5, Magdalena Alvarez Ramirez5, Francisco Diaz Cedillo6
1Green Land Landscaping and Gardening, Seedling Growth Laboratory, Brazil
2Faculty of Chemistry, California South University, USA
3University Autonomous of Campeche, Faculty of Chemical-Biological Sciences, Calle Avenida Agustin Melgar, Mexico
4Department of Marine Science, University of Calcutta, West Bengal, India
5Faculty of Nutrition, Universidad Veracruzana, Mexico
6National School of Biological Sciences of the National Polytechnic Institute, Mexico
Ricardo Gobato, Green Land Landscaping and Gardening, Seedling Growth Laboratory, 86130-000, Parana, Brazil.
Received Date: August 30, 2021; Published Date: September 29, 2021
Abstract
Proton nuclear magnetic resonance (1H NMR) is the application of nuclear magnetic resonance in NMR spectroscopy with respect to 1H within the molecules of a substance, in order to determine the structure of its molecules. The work focused on determining the 1H NMR spectrum of the molecule here called Xalapa, in homage of the city of Xalapa, the capital city of the Mexican state of Veracruz and the name of the surrounding municipality. The 1H NMR spectrum was obtained via computational methods ab initio Restricted Hartree Fock. Optimization of molecular structure via UFF, followed by PM3, RHF/EPR-II and RHF/STO-6G, thus obtaining a stable structure, in STP, NMR via the GIAO (Gauge-Independent Atomic Orbital) method. The IUPAC name of the molecule was obtained, whose composition is C: 81.7%; H: 7.1%; N: 3.4%; O: 7.8%, formula weight: 411.53536 g, and molecular formula: C28H29NO2. Limitations our study has so far been limited to computational simulation via quantum mechanics (QM) an applied theory. Our results and calculations are compatible with the theory of QM, but their physical experimental verification depends on experimental data that should be laboratory for experimental biochemical.
Keywords:Hartree-Fock method; 1H NMR spectroscopy; Xalapa; UFF; PM3; EPR-II; GIAO Methods
Introduction
The NMR (Nuclear Magnetic Resonance) spectroscopy is a spectroscopic technique to observe local magnetic fields around atomic nuclei. One magnetic field is placed in the sample and the NMR signal is produced by excitation of the nuclei sample with radio waves into nuclear magnetic resonance. The intramolecular magnetic field around an atom in a molecule changes the resonance frequency, thus giving access to details of the electronic structure of a molecule and its individual functional groups. As the fields are unique or highly characteristic to individual compounds, in modern organic chemistry practice, NMR spectroscopy is the definitive method to identify monomolecular organic compounds [1]. Proton nuclear magnetic resonance (1H NMR) is the application of nuclear magnetic resonance in NMR spectroscopy with respect to 1H within the molecules of a substance, in order to determine the structure of its molecules [2].
The work focused on determining the 1H NMR spectrum of the molecule here called Xalapa, in homage of the city of Xalapa (often spelled Jalapa) [3], the capital city of the Mexican state of Veracruz and the name of the surrounding municipality. The 1H NMR spectrum was obtained via computational methods ab initio Restricted Hartree-Fock [4-16], GIAO (Gauge-Independent Atomic Orbital) methods [17-21]. The molecule was obtained experimentally in the laboratory of the University Autonomous of Campeche (Faculty of Chemical-Biological Sciences) Valverde et al. [22-26].
Methods
Methodology
Its structure and chemical conformation and its 1H NMR spectrum were obtained from computational chemistry calculations, using the GAMESS software [15]. The methods used initially were UFF [7,12-15,27-30], obtaining the lowest energy molecular structure. Optimization of molecular structure via UFF, followed by PM3, RHF/EPR-II and RHF/STO-6G, thus obtaining a stable structure, in STP (Standard Temperature and Pressure) [7,12-15,27-30], and NMR via the GIAO method [17-21].
The properties keyword predicts NMR shielding tensors and magnetic susceptibilities using the Restricted Hartree-Fock (RHF) method, all DFT methods and the MP2 method NMR shielding tensors may be computed with the Gauge-Independent Atomic Orbital (GIAO) method [17-21]. The basis sets of Barone which are optimized for the computation of hyperfine coupling constants. EPR-II is a double zeta basis set with a single set of polarization functions.
Hartree-Fock Methods
The molecular Hartree-Fock [4-16] wave function is written as an antisymmetrized product (Slater determinant) of spin-orbitals, each spin-orbital being a product of a spatial orbital ϕi and a spin function (either α or β).
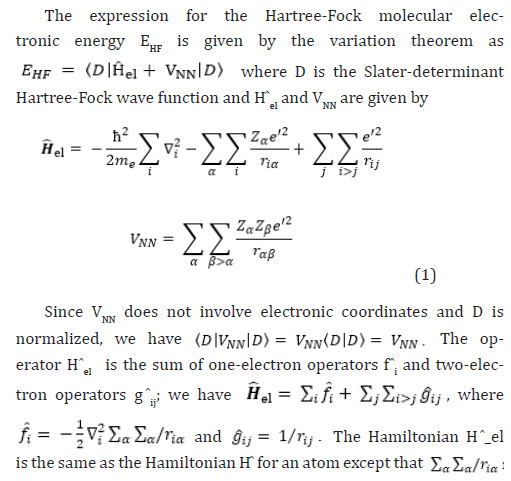
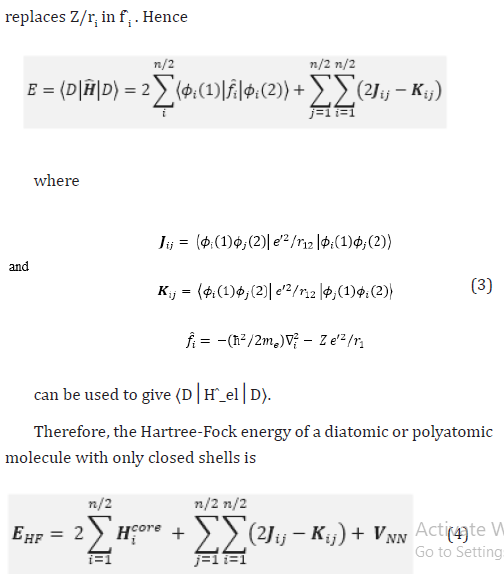
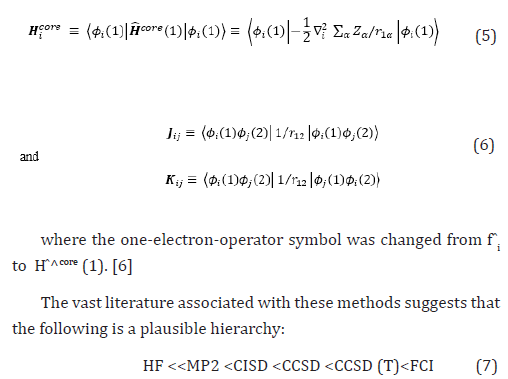
The extremes of ‘best’, FCI, and ‘worst’, HF, are irrefutable, but the intermediate methods are less clear and depend on the type of chemical problem being addressed. The use of HF [4-16] in the case of FCI was due to the computational cost.
Hardware and Software
For calculations the computer used a Desktop with SUSE Linux Enterprise Desktop [31], AMD Ryzen 7 1800X processor [32], ASUS Prime A320M-K motherboard [33], 16GB of RAM, with 500GB SSD [34]. The ab initio calculations have been performed to study the equilibrium configuration of Xalapa molecule. The set of programs GaussView 5.0.8 [35], HyperChem 8.0.6 Evaluation [36]. are the advanced semantic chemical editor, visualization, and analysis platform and GAMESS [15,35] is a computational chemistry software program and stands for General Atomic and Molecular Electronic Structure System [15,35], BIOVIA Draw 2017 [37], CHARMM22 [38] set of programs were used.
Result
The molecular structure of Xalapa molecule, were obtained through computationally calculated molecular dynamics, using the ab initio Hartree-Fock (HF) method.
The name in Figure (1) of the new molecules obtained is 2-[4-[[rac-(4S,5S,6S,10R,11S)-10-formyl-11-phenyl-5-tricyclo [7.2.0.04,6] undec-1(9)-enyl] methoxymethyl] phenyl] acetonitrile.
Xalapa molecule
Properties of Xalapa molecule:
• IUPAC name: 2-[4-[[rac-(4S,5S,6S,10R,11S)-10-formyl- 11-phenyl-5-tricyclo [7.2.0.04,6] undec-1(9)-enyl] methoxymethyl] phenyl] acetonitrile;
• PSA: 50.09;
• ALogP: 4.892;
• Stereo Center Count: 5;
• Hydrogen Acceptor Count: 3;
• Hydrogen Donor Count: 0;
• Composition: C: 81.7%; H: 7.1%; N: 3.4%; O: 7.8%;
• Formula Weight: 411.53536 g;
• Exact Mass: 411.219829178001 g;
• Molecular Formula: C28H29NO2. [37] (Figure 1-2)
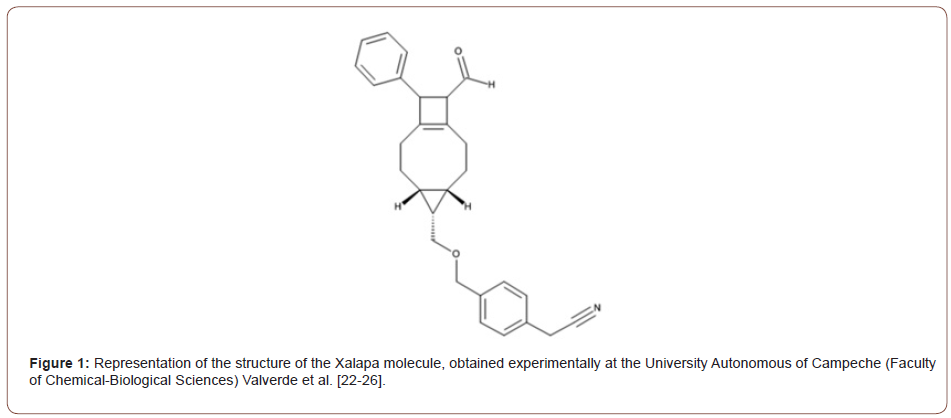
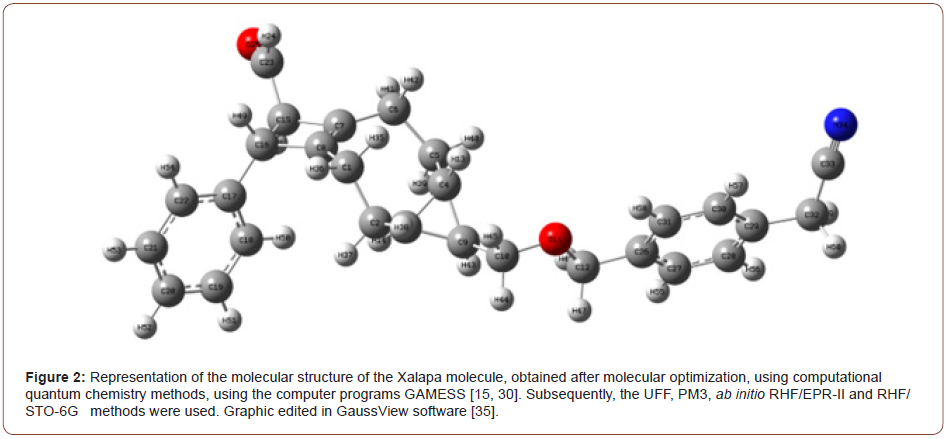
The Figure (1) representation of the structure of the Xalapa molecule, obtained experimentally at the University Autonomous of Campeche (Faculty of Chemical-Biological Sciences) Valverde et al. The Representation of the molecular structure of the Xalapa molecule, Figure (2), obtained after molecular optimization, using computational quantum chemistry methods, using the computer programs GAMESS [15, 30]. Subsequently, the UFF, PM3, ab initio RHF/EPR-II and RHF/STO-6G methods were used. Graphic edited in GaussView software.
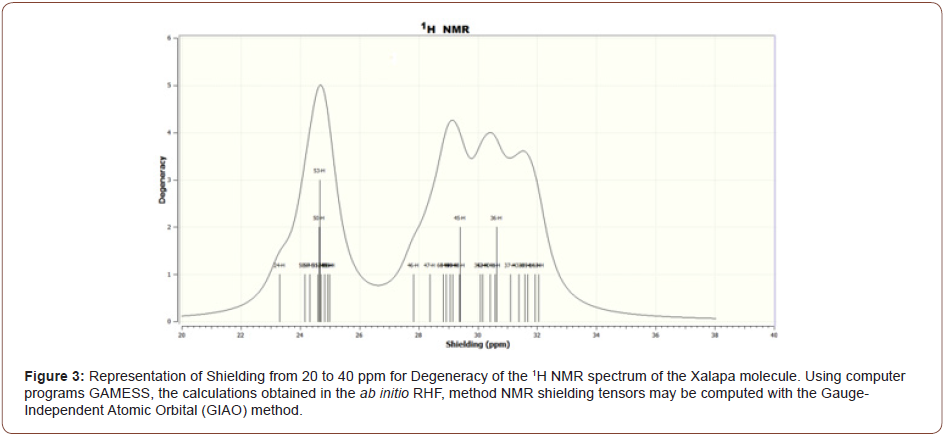
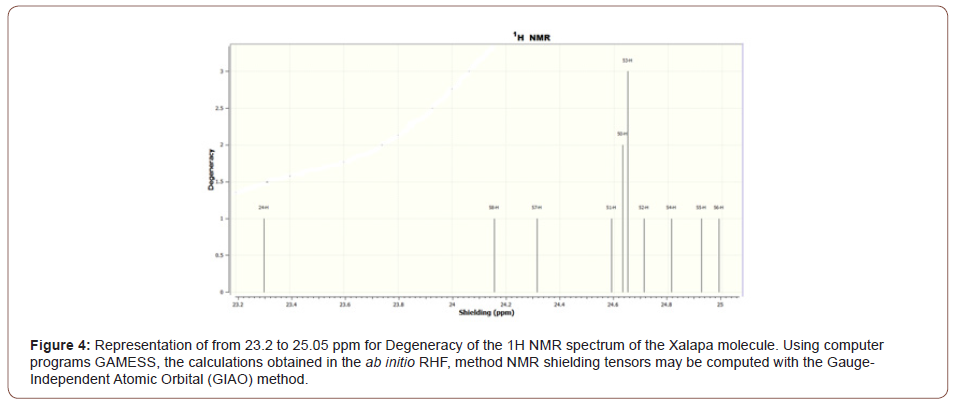
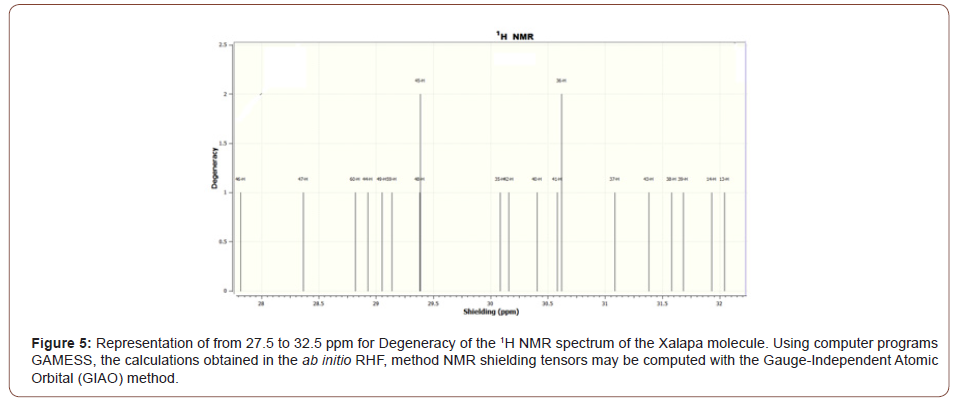
The Figure (3) representation of Shielding from 20 to 40 ppm for Degeneracy of the 1H NMR spectrum of the Xalapa molecule, The Figure (4) representation of from 23.2 to 25.05 ppm for Degeneracy of the 1H NMR spectrum of the Xalapa molecule and the Figure (5) representation of from 27.5 to 32.5 ppm for Degeneracy of the 1H NMR spectrum of the Xalapa molecule. All Using computer programs GAMESS, the calculations obtained in the ab initio RHF, method NMR shielding tensors may be computed with the Gauge-Independent Atomic Orbital (GIAO) method.
The H50, H51, and H52 hydrogens of the aromatic ring are coupling with each other to form a triplet (1:2:1), Figures (3) and (4). The H53 hydrogen is not coupling with the other H atoms and appears as a singlet, Figures (3) and (4). Hydrogen H45 from group CH2 bonded to Oxygen O11 not coupling with the other H atoms, as well as Hydrogen H36 from group CH2 from aromatic ring C9H10, bonded to Carbon C1, Figures (3) and (5).
Conclusion and challenges
The 1H NMR spectrum of the Xalapa molecule. was calculated, indicating the characteristic of the nano-molecule genesis. Characterized its 1H NMR spectrum quantum calculated, accepted by quantum chemistry parameters, with ab initio methods, in the GIAO methods. The molecule is named IUPAC 2-[4-[[rac- (4S,5S,6S,10R,11S)-10- formyl-11-phenyl-5-tricyclo [7.2.0.04,6] undec-1(9)-enyl]methoxymethyl]phenyl] acetonitrile; of composition: C: 81.7%; H: 7.1%; N: 3.4%; O: 7.8%, formula weight: 411.53536 g, and molecular formula: C28H29NO2. Limitations our study has so far been limited to computational simulation via quantum mechanics (QM) an applied theory. Our results and calculations are compatible with the theory of QM, but their physical experimental verification depends on experimental data that should be laboratory for experimental biochemical.
Acknowledgement
None.
Conflicts of Interest
No conflicts of interest.
References
- Creative Commons. (CC-BY 4.0). (2021) The Free Encyclopedia, Nuclear magnetic resonance spectroscopy.
- Proton nuclear magnetic resonance. 2021.
- Xalapa (2021). URL: https://en.wikipedia.org/wiki/Xalapa
- IN Levine (2003) Quantum Chemistry”. Pearson Education (Singapore) Pte. Ltd., Indian Branch, 482 F. IE Patparganj, 5th eedition, Delhi 110 092, India.
- A Szabo, NS Ostlund (1989) Modern Quantum Chemistry. Dover Publications, New York, USA.
- W Kohn, LJ Sham (1965) Self-consistent equations including exchange and correlation effects. Phys Rev (140): A1133.
- JM Thijssen (2001) Computational Physics. Cambridge University Press, Cambridge, USA.
- TH Dunning (1989) Gaussian basis sets for use in correlated molecular calculations. The atoms boron through neon and hydrogen J Chem Phys (ed) (90):1007–1023.
- DE Woon, TH Dunning (1993) Gaussian-basis sets for use in correlated molecular calculations. The atoms aluminum through argon J Chem Phys (98): 1358-1371.
- AK Wilson, T van Mourik, TH Dunning (1996) Gaussian basis sets for use in Correlated Molecular Calculations. Sextuple zeta correlation consistent basis sets for boron through neon J Mol Struct (Theochem) (388): 339-349.
- E Polak )1971) Computational Methods in Optimization. Fifth Avenue, pp: 77(111) New York.
- TH Dunning, PJ Hay (1977) Modern Theoretical Chemistry Plenum, New York, USA.
- E Eliav (2013) Elementary introduction to Molecular Mechanics and Dynamics.
- WJ Hehre (2003) A Guide to Molecular Mechanics and Quantum Chemical Calculations, Wavefunction, Inc, Irvine, CA, USA
- MS Gordon, MW Schmidt (2005) Advances in electronic structure theory: GAMESS a decade later. Theory and Applications of Computational Chemistry: the first forty years. Elsevier, CE Dykstra, G Frenking, K S Kim GE Scuseria (eds), pp 1167-1189, Amsterdam.
- F Weigend, R Ahlrichs (2005) Balanced basis sets of split valences, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys Chem Chem Phys (7): 3297-3305,
- F London (1937) The quantic theory of inter-atomic currents in aromatic combinations. J Phys Radium 8: 397-409.
- R McWeeny (1962) Perturbation Theory for Fock-Dirac Density Matrix Phys Rev 126: 1028.
- R Ditchfield (1974) Self-consistent perturbation theory of diamagnetism. 1. Gauge-invariant LCAO method for N.M.R. chemical shifts. Mol Phys 27: 789-807.
- K Wolinski, JF Hilton, P Pulay (1990) Efficient Implementation of the Gauge-Independent Atomic Orbital Method for NMR Chemical Shift Calculations. J Am Chem Soc 112: 8251-8260.
- JR Cheeseman, GW Trucks, TA Keith, MJ Frisch A (1996) Comparison of Models for Calculating Nuclear Magnetic Resonance Shielding Tensors. J Chem Phys 104: 5497-509.
- Lopez-Ramos Maria, Figueroa-Valverde Lauro, Diaz-Cedillo Francisco, Gobato Ricardo, Heidari Alireza, et al. (2021) Synthesis of two 2,3,4,5-tetrahydrooxepin-7-ylamino) benzoate derivatives as antibacterial agents against Escherichia coli and Staphylococcus aure. Vietnam Journal of Chemistry. Wiley in press.
- M Lopez-Ramos, L Figueroa-Valverde, F Diaz-Cedillo, M Rosas-Nexticapa, MV Mateu-Armand, et al. (2018) Facile synthesis of some oxazepine- steroid using three component system, Parana J Sci Educ 6: 34-45.
- L Figueroa-Valverde, F Diaz-Cedillo, M Lopez-Ramos, M, Rosas-Nexticapa, V Mateu-Armad, et al. (2019) Experimental and theoretical evaluation of two indol-steroid-cyclobuta-imidazole derivatives as antibacterial drugs, Biointerface Res Appl Chem 9(5): 4405-4415.
- L Figueroa-Valverde, F Diaz-Cedillo, A Camacho-Luis (2010) A Synthesis of a succinate– dihydrotestosterone–dihydropyrimidine conjugate, Monats. Fur Chem 141(1): 75-78.
- L Figueroa-Valverde, M Rosas-Nexticapa, M Lopez-Ramos, Diaz-cedillo (2020) Synthesis of a New Dioxaspiro [bicyclo [3.3. 1] nonane-oxabicyclo [6.2. 0] deca-1 (10), 8-dien-4-one Derivative Using Some Chemical Strategies. Lett Org Chem17(5): 393-402.
- R Gobato, MRR Gobato, A Heidari (2019) Raman Spectroscopy Study of the Nano Molecule C13H20BeLi2SeSi Using ab initio and Hartree–Fock Methods in the Basis Set CC–pVTZ and 6–311G** (3df, 3pd)”, International Journal of Advanced Engineering and Science 7(1): 14–35,
- R Gobato, A Heidari (2018) Infrared Spectrum and Sites of Action of Sanguinarine by Molecular Mechanics and ab initio Methods. International Journal of Atmospheric and Oceanic Sciences 2(1): 1-9.
- A Heidari, R Gobato (2020) Investigation of the internal structure and dynamics of gum cancer cells, tissues and tumors by 13C–NMR spectra of DNA/RNA of gum cancer cells as an essential structural tool for integrative studies of gum cancer cells development. Dent Oral Maxillofac Res 6: 1-3
- MS Gordon et al. (1993) General atomic and molecular electronic structure system (GAMESS). J Comput Chem 14: 1347-1363,
- Suse (2021) SUSE Linux Enterprise Desktop.
- (2021) AMD Advanced Micro Devices (2021), Inc Processador AMD Ryzen™ 7 1800x.
- (2021) ASUS. AMD AM4 uATX motherboard with LED lighting, DDR4 3200MHz, 32Gb/s M.2, HDMI, SATA 6Gb/s, USB 3.0.
- (2021) Creative Commons. (CC-BY 4.0). Wikipedia, The Free Encyclopedia, Solid-state drive.
- (2009) R Dennington, T Keith, J Millam. Gaussview.
- Molecular Modeling System. Hyper Chem TM 7.5 evaluation (2003) Hypercube.
- BIOVIA Draw 2017 Enterprise. MDL Draw Editor 17.1.0.900 (2017) Computational results obtained using software programs from Dassault Systemes BIOVIA. The ab initio calculations were performed with the DMol3 program, and graphical displays generated with Draw.
- R Brooks, RE Bruccoleri, BD Olafson, DJ States, S Swaminathan, et al. (1983) CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations J Comp Chem 4: 187-217.
- BR Brooks, C L Brooks, AD MacKerell, L Nilsson, RJ Petrella, et al. (2009) CHARMM: The Biomolecular Simulation Program. J Comput Chem 30: 1545-1614.
-
Ricardo Gobato, Alireza Heidari, Lauro Figueroa Valverde, Abhijit Mitra, Marcela Rosas Nexticapa etc al.. 1H NMR Spectroscopy of the New Xalapa Molecule. Arch Biomed Eng & Biotechnol. 6(1): 2021. ABEB.MS.ID.000627.
-
Hartree-Fock method, 1H NMR spectroscopy, Xalapa, UFF, PM3, EPR-II, GIAO Methods, Gauge-Independent Atomic Orbital, Optimization, Molecular structure
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






