 Opinion
Opinion
Improved Algorithm of Blocking the Selected Edges in the Digraph
Tsitsiashvili Gurami, Institute for Applied Mathematics FEB RAS, Vladivostok, Russia.
Received Date: October 02, 2019; Published Date: October 11, 2019
Abstract
In this paper, a protein network represented by a directed graph is considered. The problem of determining the minimum number of edges that break paths from the input proteins of the network to the output ones and passing through some subset of proteins in this network is analyzing. An improved algorithm based on a selection of connectivity components in the sub-graph with dedicated subset of nodes is suggesting.
Keywords: Cluster; Digraph; Sub-graph; Protein network; Connectivity component
Introduction
In this paper, we consider a protein network represented by a directed graph (digraph) G with the set U of nodes, which are proteins and whose directed edges are paired bonds between nodes represented in the Cytoscape program. Dedicate the subset U/ ⊂U of nodes and decrease in a comparison with [1] a number of edges, which block all paths from outside of the set U/ outside of the set U/ .
Take all nodes from the subset U/ and all edges between them.
These nodes and edges create directed sub-graph G/ ⊂ G. Replace
all (directed) edges of the sub-graph G/ ⊂ G by undirected ones
and obtain undirected graph G// . Define in the sub-graph G// all its
connectivity components 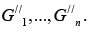 .Return directions to all edges
of the sub-graph G//K and define in such a way the sub-graph k G of
the digraph G, k =1,...,n.
.Return directions to all edges
of the sub-graph G//K and define in such a way the sub-graph k G of
the digraph G, k =1,...,n.
Factorize each sub-graph Gk by a relation of cyclic equivalence
and construct acyclic digraph with nodes are clusters of cyclic
equivalence. In the sub-graph, Gk each cluster has out coming edges
to another cluster and/or incoming edges from another cluster. If a
cluster has only out coming edges to another cluster, we call it input
cluster. If a cluster has only incoming edges from another cluster,
we call it output cluster. In the sub-graph, Gk there is a path from
input cluster to some of output clusters and there is a path to output
cluster from some input ones. Any edge beginning in the sub-graph
Gk does not reach another sub-graph 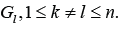
Denote by 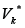 the set of edges incoming to GK and by
the set of edges incoming to GK and by 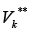 the
set of edges out coming from .GK It is obvious that
the
set of edges out coming from .GK It is obvious that
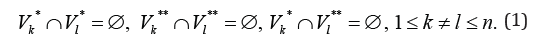
For any edge 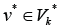 there is a path to some edge
there is a path to some edge 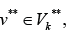 and for any edge
and for any edge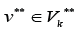 there is a path from some edge
there is a path from some edge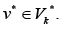 Formula (1) allows to consider separately incoming and out coming edges of the sub-graphs
Formula (1) allows to consider separately incoming and out coming edges of the sub-graphs 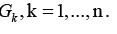
Designate by 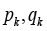 numbers of edges in the sets
numbers of edges in the sets 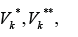 .relatively. Assume that
.relatively. Assume that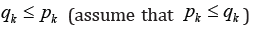 and block
all edges from
and block
all edges from 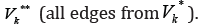 In such a way, we block all
paths from the outside of Gk outside of .Gk Then a number of edges
blocking the sub-graph k Gk equals
In such a way, we block all
paths from the outside of Gk outside of .Gk Then a number of edges
blocking the sub-graph k Gk equals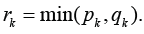 Consequently
to block the set U/ of dedicated nodes it is necessary only to block
the following number of edges
Consequently
to block the set U/ of dedicated nodes it is necessary only to block
the following number of edges
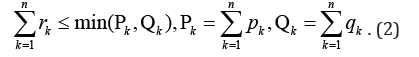
Formula (2) confirms that suggested algorithm based on the definition of connectivity components in the sub-graph G// decreases a number of blocked edges in a comparison with [1].
Acknowledgement
None.
Conflict of Interest
No conflict of interest.
-
Tsitsiashvili Gurami. Improved Algorithm of Blocking the Selected Edges in the Digraph. Annal Biostat & Biomed Appli. 3(3): 2019. ABBA.MS.ID.000561.
Schrödinger equation; Variable; Wave function; Energy, Density, Energy of Light, Wave Function, Final Time; Own Operator; Volume Energy; Speed of Light; Function of the Function Variable of Wave.
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






