 Conceptual Paper
Conceptual Paper
Decomposition Algorithms of Blocking the Selected Edges in the Digraph
Tsitsiashvili Gurami, Institute for Applied Mathematics FEB RAS, Vladivostok, Russia. .
Received Date: October 17, 2019; Published Date: October 22, 2019
Abstract
In this paper, a protein network represented by a directed graph is considered. The problem of determining the minimum number of edges that break paths from the input proteins of the network to the output ones and passing through some subset of proteins in this network is analyzing. A decomposition algorithms are basing on a selection of classes of cyclically equivalent nodes in the sub-graphs with dedicated subset of nodes is suggesting.
Keywords: Cluster; Digraph; Sub-graph; Protein network; Connectivity component
Introduction
By analogy with [1], we consider a protein network represented by a directed graph (digraph) G with the set U of nodes, which are proteins and whose directed edges are paired bonds between nodes represented in the Cytoscape program. Dedicate the subset U/ ⊆U of nodes and decrease in a comparison with [1] a number of edges, which block all paths from outside of the set U/ outside of the setU/ .
Take all nodes from the subset U/ and all edges between them. These nodes and edges create directed sub-graphG/ ⊆ G . Replace all (directed) edges of the sub-graph G/ by undirected ones and obtain undirected graph G// . Define in the sub-graph G// all its connectivity components / / / / Return directions to all edges of the subgraph G//k G k and define in such a way the sub-graph Gk of the digraph G, k =1,...,n.
Factorize each sub-graph Gk by a relation of cyclic equivalence (two nodes are cyclically equivalent if there is a cycle containing both of them) and construct acyclic digraph with nodes - clusters of cyclic equivalence. Construct the partial order ≥ between the clusters (a relation A ≥ B is true if there is a way from cluster A to cluster B ) in Gk .
Denote by vk *a set of edges incoming to GK and by VK**a set
of edges out-coming from GK and designate bypK, qKnumbers of
edges in these sets. Define the following sets WK*,WK**of clusters
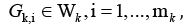 in digraphGK Each cluster
in digraphGK Each cluster
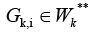 has
final nodes of some edges from the set V
K*Similarly, each cluster
has
final nodes of some edges from the set V
K*Similarly, each cluster 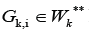 has initial nodes of some edges from the set VK
**The WK*,WK** setsmay intersect.
has initial nodes of some edges from the set VK
**The WK*,WK** setsmay intersect.
Construct the set 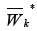 consisting of clusters GK,i,such that
consisting of clusters GK,i,such that
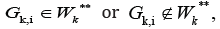 but there are some cluster
but there are some cluster
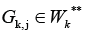 and a way from
and a way from
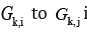 in digraph GK.By analogy construct the set
in digraph GK.By analogy construct the set
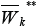 of clusters
of clusters
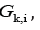 such that
such that
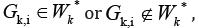 but
there are some cluster
but
there are some cluster 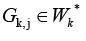 and a way from
and a way from
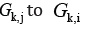 in
digraph GK
in
digraph GK
Designate by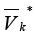 all edges incoming some clusters containing
in the set
all edges incoming some clusters containing
in the set 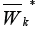 from the outside of the graph
from the outside of the graph
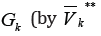 all edges
out-coming from some clusters in the set WK**outside the graph GKDenote
all edges
out-coming from some clusters in the set WK**outside the graph GKDenote
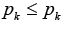 a number of edges in the set
a number of edges in the set
 a number of edges in the set
a number of edges in the set
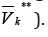
Assume that 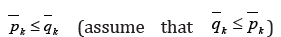 and block
all edges from
and block
all edges from  In such a way,
we block all paths from the outside of GKoutsideGK. Then a number of edges blocking the sub-graph GKequals’
In such a way,
we block all paths from the outside of GKoutsideGK. Then a number of edges blocking the sub-graph GKequals’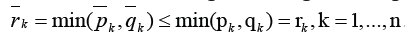 Last inequalities
show, how this algorithm decreases a number of blocked edges in a comparison with [1]. Next step of the decomposition procedure in
this algorithm may be an allocation of disconnected subsets in pairs
of sets
Last inequalities
show, how this algorithm decreases a number of blocked edges in a comparison with [1]. Next step of the decomposition procedure in
this algorithm may be an allocation of disconnected subsets in pairs
of sets 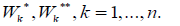
Acknowledgement
Supported by Russian Fund of Basic Research’s, project 17-07- 00177.
Conflict of Interest
No conflict of interest.
References
- G Sh Tsitsiashvili (2019) Improved algorithm of blocking the selected nodes in the digraph, Annals of Biostatistics and Biometric Applications 3(3).
-
Tsitsiashvili Gurami. Decomposition Algorithms of Blocking the Selected Edges in the Digraph. Annal Biostat & Biomed Appli. 3(3): 2019. ABBA.MS.ID.000562.
Decomposition Algorithms, Selected Edges, Digraph, Cluster; Digraph; Sub-graph; Protein network; Connectivity component, Network, Analyzing, Applied Mathematics,
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






