 Research Article
Research Article
Fitting of a multivariable nonlinear data set: A suggested strategy in lactic fermentations
Francisco Caballero*
Carrera de Ingeniería Química. Facultad de Estudios Superiores Zaragoza UNAM
Francisco Caballero, Carrera de Ingeniería Química. Facultad de Estudios Superiores Zaragoza UNAM, Mexico.
Received Date:March 02, 2023; Published Date:March 27, 2023
Abstract
Nowadays modeling in bioengineering process is mature field, but experimental data fitting is procedure that keeps on practice, the model obtained can guarantee control and process modelling applications. In this work a set of multivariable nonlinear data, that comes from lactic fermentation by lactobacillus acidophilus. In this way, the fitting procedure is achieved by following different theoretical tips. The data was taken from Escorza [1] to obtain the parameters and they were compared with another kind of lactobacillus [2]. The procedure suggested, can be used to fitting different biotechnology data with precision and confidence.
Keywords:Non-linear fitting; Time series data; Lactic acid; Levenberg-Marquardt Method
Introduction
Lactic acid is substance very important in actual market due to different products of aggregated high value. In this context and biotechnological route preferred because its specificity i.e., is possible produce only Levo (L) optical isomer an important material to produce poly acid lactic, ethyl lactate and different specialties [3].
Inside the most commons microorganism used to lactic fermentation Lactobacillus acidophilus have highlighted and another different species as L. dembruki, L. rhamnosus and so on [4].
Procedure description
In spite different methodologies to get parameters. In this
time, it is going to apply the Levenberg Marquardt Method based
in biotechnological data [5], but under little pieces of theorical
fundamentals and one that other suggestion. This is main scope of
this work.
1. This is the main idea: every kind model can be used to
represent the behavior of different phenomena. However, it must
be capable to reproduce the experimental behavior under the
following establishment:

Where experimental data yiE going to be represented by a model yi(p,xi) that depends on different variables xi (i e. multivariate for this vectorial notation) under a set of physical parameters p (not necessarily). The later expression defines the square error (ε ).
2. Under later scheme, it must be obtained the set of first partial derivatives ∂ε/∂pi; where the parameter i p belongs a set of p . So that, there are many parameters as the range of vector p .
3. The set of ∂ε/∂pi=0 is the first derivative and is possibly get the minimum square error. In this time, it has a set of nonlinear algebraic equations and maybe you are disappointed, because no new today. However, if they are established the equations, it recognizes that, it is not trivial procedure, it must keep on mind that all the equations contain a summation of all experimental data.
Until this step, it seems that does not appear some difficulty; but be careful¡ Let’s see the following nonlinear expression:

4. Analyzing it is possible to observes that x (biomass L. Acidophilus), S (substrate i.e., glucose) and P (product in this case, lactic acid) are the independent variables and the change rate of biomass (dx/dt ) is the dependent variable. By analyzing (Figure 1a) y’s left side are plotting the concentration but no the derivatives as eq. (2) requires. Then is necessary get the derivatives and in this way, it will be possible to satisfy eq. 2. This the reason that in y’s right side are plotting the derivative of concentrations in figure 1a. So, it must choice green scatters of right axe. By substituting experimental data derivatives (equivalent to yiE ) and independent variables (x, S and P; right side eq. 2) into eq. (1) becomes:

5. So, the minimal is obtained. The procedure gets μ max ,Ks,Ki,kd and keeps the values in order to solve the rest of parameters, because of the full model is represented by:

By looking at eq. 3, only it will be solved mmax , and Yx/s (Remember the values of μmax,Ks,Ki,kd were obtained already, only you must substitute them), and is possible simplify eq. 3 with the following relationship Yx/s =1/Ys/x and procedure becomes in a linear fitting. By fitting eq. (3) red scatters on right side Fig. 1a were used. Finally, repeat the procedure in eq. 4 by using blue scatters on right side Figure 1a.

As can be seen, the process is systematic and an adjustment
is made by equation, and by keeping the parameters that were
obtained earlier, each time the adjustment is easier and can be
compared according to field of study.
Discussion
Table 1:Parameters obtained by nonlinear fitting.
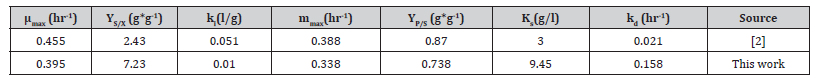
Table 1 shows the value of parameters done by this procedure and are contrasted with results reported [2] is possible evaluate about the values cause are the same magnitude and is possible because data of this work comes from lactic fermentation by lactobacillus acidophils while data from [2] is the metabolism of lactobacillus casei subsp. rhamnosus.
However, one of the bests tests is simulating the set 2-4 with parameter obtained; in this occasion, simulation is performed in solid thinking Embed 2017.1 (formerly Vissim), results can see it in figures 1.b where the continuous line is the simulation of the model in accordance with experimental scatter color respectively. In a global vision, the result of fitting procedure successfully represents fermentation phenomena.
To close this work based on a theoretical support, very known, but with analytic and deep vission, is important to mention data of this work comes from a batch reactor. Therefore, it was possible to get a total biokinetics, later, the parameters can be fed to establish conditions in different reactors as fed batch reactor or a continuous reactor, very common kind reactors in biotechnological field.
Conclusion
By solving the parameters equation by equation is a theoretical procedure that let to user evaluate and compare it different models and tunning the best alternative without need high computation, further the method is simplify instead to solve full vector what might it require constrained nonlinear programming, that, it could be the next step. Finally, always is recommendable have parameters reported by another investigators (in this work the set of eq. 2-4 were taken and suggested by [2] under the theoretical discussion in [6]) to calibrate the own procedure made under these recommendations.
By the way the derivatives of time data series can be obtained by polynomial fitting but don’t confuse yourself the purpose is get the derivatives to polynomial mentioned and use as dependent variables as can see in first term of summation in eq. 2a. Finally, with all data required, is possible use numerical procedures or use of software. In this case Mathcad was used to solve eq. 2a under its SSE procedure.
Acknowledgement
To DGAPA UNAM for financial support under PAPIIT project No. 112122
Conflict of Interest
None.
References
- Escorza V G, Membrillo Venegas I, Cruz Diaz M, Arcos Casarrubias J A, Caballero F, et al. (2014) Determination of kinetic parameters to produce lactic acid by Lactobacillus acidophilus (In spanish). CIM Journal 2(1): 619-625.
- Lombardi M, Fiaty K, Laurent P (1999) Implementation of observer for on-line estimation of concentration in continuous-stirred membrane bioreactor: Application to the fermentation of lactose. Chemical Engineering Science 54: 2689-2696.
- Jiang X, Luo Y, Tian X, Huang D, Reddy N, et al. (2010) Chemical structure of poly(lactic acid). In R. Auras, L-T. Lim, S. E. M. Selke & H. Tsuji, Poly (lactic acid) Synthesis, Structures, Properties, Processing, and Applications (69-82). Wiley.
- Mora Villalobos JA, Montero Zamora J, Barboza N, Rojas Garbanzo C, Usaga J, et al. (2020) Multi-Product Lactic Acid Bacteria Fermentations: A Review. Fermentation 6(1): 23.
- Dorantes Landa D N, Cocotle Ronzón Y, Morales Cabrera M A, Hernández Martínez E (2020) Modelling of the xylitol production from sugarcane bagasse by immobilized cells. Journal of Chemical Technology & Biotechnology 95: 1936-1945.
- Bouguettoucha A, Balannec B, Amrane A (2011) Unstructured Models for Lactic Acid Fermentation-A Review. Food Technology and Biotechnology 49(1): 3-12.
-
Francisco Caballero*. Fitting of a Multivariable Nonlinear Data Set: A Suggested Strategy in Lactic Fermentations. Annal Biostat & Biomed Appli. 5(2): 2023. ABBA.MS.ID.000606.
Non-linear fitting, Time series data, Lactic acid, Levenberg marquardt method, Nonlinear programming, Fermentation phenomena, Nonlinear algebraic equations, Variables
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






