 Case report
Case report
Human Intestinal Microbiota in Response to Antibiotics, Nicotine and Caffeine
Martynova Van Kley Alexandra*, Rodriguez Maximiliano and Rorex Colin B
Department of Biology, Stephen F. Austin State University, Nacogdoches, TX, USA
Jezdimir Knezevic, Mirce Akademy, Woodbury Park, Exeter, UK.
Received Date:September 23, 2019; Published Date:September 27, 2019
Abstract
Intestinal microbiota play a number of critical roles in the health of the organism. They assist in nutrient metabolism and play a key role in the host’s immune response. Current research is revealing that there may be a significant impact from intestinal bacteria on the gut-brain axis and that they play a downstream role in several neurological disorders.
Case presentation: This pilot study is looking at the effects of antibiotic use on a healthy 21 years old male’s intestinal microbiota during a course of amoxicillin usage and the recovery period after treatment was completed.
Keywords:Gut microflora; Antibiotics; Nicotine; Caffeine; Lack of sleep
Background
Previous work shows that intestinal microbiota varies significantly from individual to individual with changes in diet, exercise, smoking and a myriad of other factors resulting in changes to microbial diversity. Thus, fundamental to development of any therapeutic intestinal microbiota treatments, we need to have a mechanistic understanding of how bacterial diversity are impacted by these factors. The goal of this project was to see how behavioral factors influence both the antibiotic driving decline of gut microbiota but also the recovery period following an antibiotic course.
Case Presentation
Stool sample collection was conducted over 26 days with 16 sample points along with detailed diet description was gathered. DNA extraction from all sixteen samples was done by an automated Maxwell16 system using a Maxwell 16 Tissue DNA purification kit. Several methods were then performed to analyze the microbial community. First, microbial populations were compared by examining differences in G/C content of PCR products following amplification with primers for the high variable V3 region of the 16S ribosome gene. PRC products were visualized using Denaturing Gradient Gel Electrophoresis (DGGE) which allows for separation based on annealing strength of DNA fragments. Dendrogram band pattern relatedness from DGGE was analyzed using GelCompare II, v6.6 11 (Applied Maths, Austin, TX) based on the Dice similarity coefficient and the unweighted pair group method using arithmetic averages (UPGMA) for cluster analysis and comparison according to percentage similarity coefficient (%SC). Extracted DNA samples were also sent for outside sequencing (Research and Testing Laboratories, Lubbok, TX). Samples were amplified using a primer covering the V3 and V4 regions of the 16S ribosome gene and sequenced on the Illumina platform. The sequences were then submitted to the RDP classifier to identify bacterial species. A taxonomic matrix was assembled using an 80% cut-off threshold to the lowest possible taxonomic level. This was analyzed using non-metric multidimensional scaling (NMDS) in R Studio (R Studio, Boston, MA).
As expected NMDS data (Fig.1) indicates that the microbiome for the 16 samples is split into two groups based on the antibiotic course [1]. Sample 1 was collected on the same day as the antibiotic regimen began, before there would be time for antibiotics to accumulate and impact the intestinal microbiota. S2, collected 24 hrs. after S1 shows how rapidly the microbiome changes in response to antibiotics. Likewise, S9 and S10 are also transition points showing how rapidly discontinuation of antibiotic treatment results in changes of species composition (Figure 1).
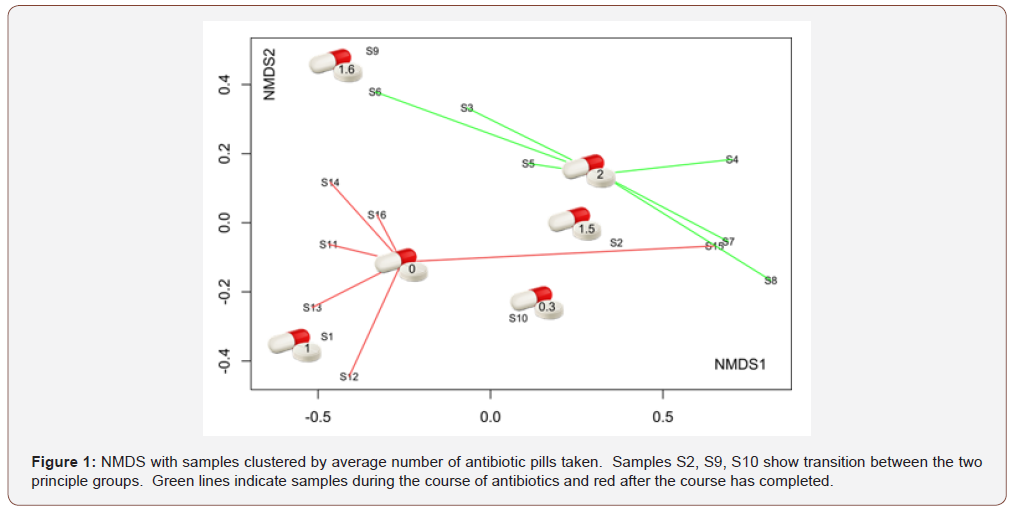
An anomaly here is sample S15 which is unexpectedly grouping with the antibiotic-treated samples and not with the untreated samples. The prior samples (S13 and S14) aren’t showing any shift like the transition points S9 and S10. We suspect that the S15 is being driven by a lack of sleep. The day prior to collection of S15 the subject reported only 4 hours of sleep and lack of sleep has been reported to have an effect on the gut microbiota [2].
Discussion
We found that the antibiotic treatment had the strongest effect on the microbiome community with samples collected during the course of antibiotics grouping together in ordination space and samples collected after the course of antibiotics had completed also grouping together.
Acknowledgement
None.
Conflict of Interest
No Conflicts of Interest.
References
- Pérez Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, et al. (2013) Gut microbiota disturbance during antibiotic therapy: a multiomic approach. Gut (11): 1591-1601
- Benedict C, Vogel H, Jonas W, Woting A, Blaut M, et al. (2016) Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Molecular Metabolism 5(12): 1175–118
-
Martynova Van Kley Alexandra, Rodriguez Maximiliano, Rorex Colin B. Human Intestinal Microbiota in Response to Antibiotics, Nicotine and Caffeine. Arch Biomed Eng & Biotechnol. 2(5): 2019. ABEB.MS.ID.000550.
-
Gut microflora, Antibiotics, Nicotine, Caffeine, Lack of sleep, Microbial community, Human Intestinal Microbiota, Antibiotic treatment,
-

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.






